Development of a p62 biodegrader for autophagy targeted degradation.
Thiel, Z., Marcellin, D., Manneville, C., Goretzki, B., Egger, L., Maher, R., Siccardi, N., Torres, L., Probst, A., Muller, C.S., George, N., Vogel, M., Sinterhauf, S., Lavoisier, A., Choi, J.Y., Forcellino, L., Landshammer, A., Hauck, P., Be, C., Villard, F., Gutmann, S., Meyer, M., Freuler, F., Hinniger, A., Fernandez, C., Chau, S., Patoor, M., Sansig, G., Mitchell, G., Nyfeler, B.(2025) Nat Commun 16: 10858-10858
- PubMed: 41339312
- DOI: https://doi.org/10.1038/s41467-025-65868-9
- Primary Citation of Related Structures:
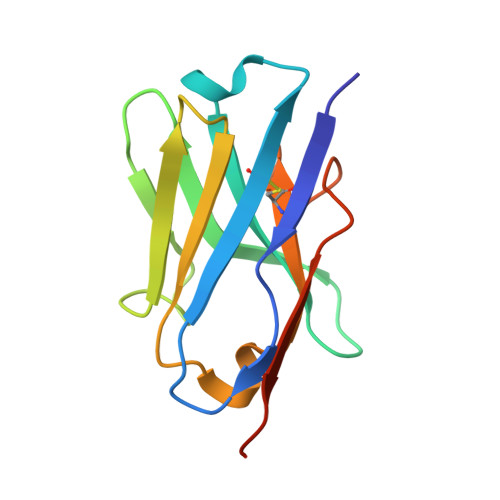
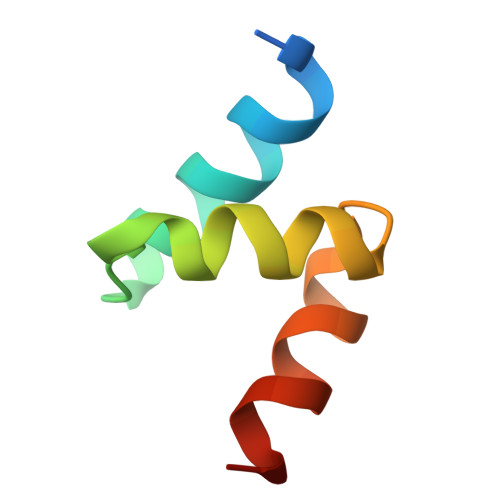
9H1J - PubMed Abstract:
Autophagy-based targeted degradation offers a powerful complement to proteasomal degradation leveraging the capacity and versatility of lysosomes to degrade complex cargo. However, it remains unclear which components of the autophagy-lysosomal pathway are most effective for targeted degradation. Here, we describe two orthogonal induced-proximity strategies to identify autophagy effectors capable of degrading organelles and soluble targets. Recruitment of autophagy cargo receptors, ATG8-like proteins, or the kinases ULK1 and TBK1 is sufficient to trigger mitophagy, while only autophagy cargo receptors capable of self-oligomerization degrade soluble cytosolic proteins. We further report a single-domain antibody against p62 and its use as a heterobifunctional degrader to clear mitochondria. Fusing the p62 single-domain antibody to PINK1 enables selective targeting of damaged mitochondria. Our study highlights the importance of avidity for targeted autophagy and suggests that autophagy cargo receptors are attractive entry points for the development of heterobifunctional degraders for organelles or protein aggregates.
- Discovery Sciences, Biomedical Research, Novartis, Basel, Switzerland. zacharias.thiel@novartis.com.
Organizational Affiliation: