Universal PSII assembly intermediate
Fadeeva, M., Klaiman, D., Nelson, N.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Currently 9GNW does not have a validation slider image.
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II protein D1 | 336 | Dunaliella salina | Mutation(s): 0 EC: 1.10.3.9 Membrane Entity: Yes | 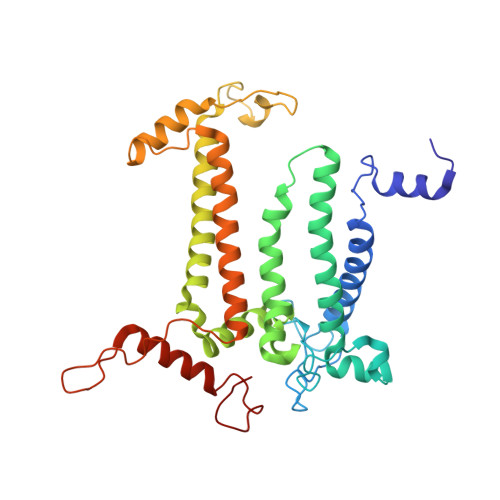 | |
UniProt | |||||
Find proteins for D0FY08 (Dunaliella salina) Explore D0FY08 Go to UniProtKB: D0FY08 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | D0FY08 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II CP47 reaction center protein | 484 | Dunaliella salina | Mutation(s): 0 Membrane Entity: Yes | 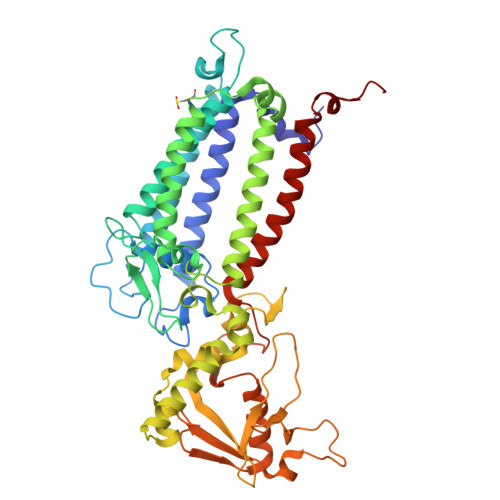 | |
UniProt | |||||
Find proteins for A0A1C8XRM6 (Dunaliella salina) Explore A0A1C8XRM6 Go to UniProtKB: A0A1C8XRM6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1C8XRM6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein Psb30 | C [auth V] | 33 | Dunaliella salina | Mutation(s): 0 Membrane Entity: Yes | 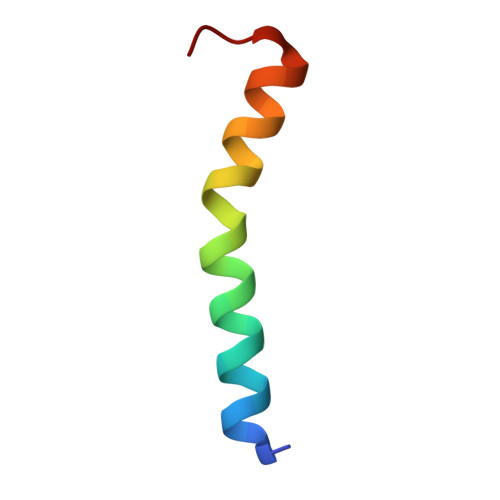 |
UniProt | |||||
Find proteins for D0FY13 (Dunaliella salina) Explore D0FY13 Go to UniProtKB: D0FY13 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | D0FY13 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II CP43 reaction center protein | D [auth C] | 449 | Dunaliella salina | Mutation(s): 0 Membrane Entity: Yes | 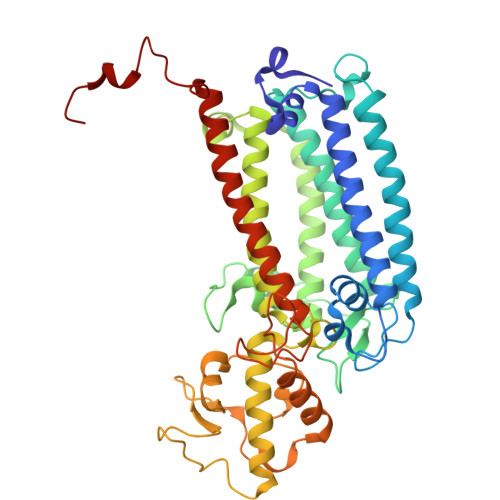 |
UniProt | |||||
Find proteins for D0FXY3 (Dunaliella salina) Explore D0FXY3 Go to UniProtKB: D0FXY3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | D0FXY3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II D2 protein | E [auth D] | 348 | Dunaliella salina | Mutation(s): 0 EC: 1.10.3.9 Membrane Entity: Yes |  |
UniProt | |||||
Find proteins for A0A1C8XRK9 (Dunaliella salina) Explore A0A1C8XRK9 Go to UniProtKB: A0A1C8XRK9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1C8XRK9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome b559 subunit alpha | F [auth E] | 76 | Dunaliella salina | Mutation(s): 0 Membrane Entity: Yes |  |
UniProt | |||||
Find proteins for D0FY01 (Dunaliella salina) Explore D0FY01 Go to UniProtKB: D0FY01 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | D0FY01 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome b559 subunit beta | G [auth F] | 31 | Dunaliella salina | Mutation(s): 0 Membrane Entity: Yes |  |
UniProt | |||||
Find proteins for A0A1C8XRP4 (Dunaliella salina) Explore A0A1C8XRP4 Go to UniProtKB: A0A1C8XRP4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1C8XRP4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 8 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein H | 67 | Dunaliella salina | Mutation(s): 0 Membrane Entity: Yes |  | |
UniProt | |||||
Find proteins for A0A1C8XRP3 (Dunaliella salina) Explore A0A1C8XRP3 Go to UniProtKB: A0A1C8XRP3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1C8XRP3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 9 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein I | 33 | Dunaliella salina | Mutation(s): 0 Membrane Entity: Yes |  | |
UniProt | |||||
Find proteins for D0FXX5 (Dunaliella salina) Explore D0FXX5 Go to UniProtKB: D0FXX5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | D0FXX5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 10 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein J | 36 | Dunaliella salina | Mutation(s): 0 Membrane Entity: Yes |  | |
UniProt | |||||
Find proteins for A0A1C8XRM8 (Dunaliella salina) Explore A0A1C8XRM8 Go to UniProtKB: A0A1C8XRM8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1C8XRM8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 11 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein K | 37 | Dunaliella salina | Mutation(s): 0 Membrane Entity: Yes |  | |
UniProt | |||||
Find proteins for D0FXX2 (Dunaliella salina) Explore D0FXX2 Go to UniProtKB: D0FXX2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | D0FXX2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 12 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein L | 36 | Dunaliella salina | Mutation(s): 0 Membrane Entity: Yes |  | |
UniProt | |||||
Find proteins for D0FY19 (Dunaliella salina) Explore D0FY19 Go to UniProtKB: D0FY19 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | D0FY19 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 13 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein M | 31 | Dunaliella salina | Mutation(s): 0 Membrane Entity: Yes |  | |
UniProt | |||||
Find proteins for D0FXZ3 (Dunaliella salina) Explore D0FXZ3 Go to UniProtKB: D0FXZ3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | D0FXZ3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 14 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| PsbO | N [auth O] | 238 | Dunaliella salina | Mutation(s): 0 Membrane Entity: Yes |  |
UniProt | |||||
Find proteins for A0A7S3QTM3 (Dunaliella tertiolecta) Explore A0A7S3QTM3 Go to UniProtKB: A0A7S3QTM3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A7S3QTM3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 15 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| PsbP | O [auth P] | 187 | Dunaliella salina | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A9J4RF14 (Dunaliella salina) Explore A0A9J4RF14 Go to UniProtKB: A0A9J4RF14 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A9J4RF14 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 16 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein T | P [auth T] | 30 | Dunaliella salina | Mutation(s): 0 Membrane Entity: Yes |  |
UniProt | |||||
Find proteins for D0FY04 (Dunaliella salina) Explore D0FY04 Go to UniProtKB: D0FY04 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | D0FY04 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 17 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| PSII 6.1 kDa protein | Q [auth W] | 45 | Dunaliella salina | Mutation(s): 0 Membrane Entity: Yes |  |
UniProt | |||||
Find proteins for A0A9J4RF15 (Dunaliella salina) Explore A0A9J4RF15 Go to UniProtKB: A0A9J4RF15 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A9J4RF15 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 18 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II PsbX | R [auth X] | 33 | Dunaliella salina | Mutation(s): 0 Membrane Entity: Yes |  |
UniProt | |||||
Find proteins for A0A7S3VKF3 (Dunaliella tertiolecta) Explore A0A7S3VKF3 Go to UniProtKB: A0A7S3VKF3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A7S3VKF3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 19 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein Z | S [auth Z] | 61 | Dunaliella salina | Mutation(s): 0 Membrane Entity: Yes |  |
UniProt | |||||
Find proteins for D0FXZ2 (Dunaliella salina) Explore D0FXZ2 Go to UniProtKB: D0FXZ2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | D0FXZ2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 20 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein U, PsbU | T [auth U] | 28 | Dunaliella salina | Mutation(s): 0 |  |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 19 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| DGD Query on DGD | DB [auth B], XB [auth C], YB [auth C], ZB [auth C] | DIGALACTOSYL DIACYL GLYCEROL (DGDG) C51 H96 O15 LDQFLSUQYHBXSX-HXXRYREZSA-N |  | ||
| CLA Query on CLA | BA [auth A] CC [auth D] EC [auth D] FC [auth D] HB [auth C] | CHLOROPHYLL A C55 H72 Mg N4 O5 ATNHDLDRLWWWCB-AENOIHSZSA-M |  | ||
| PHO Query on PHO | AA [auth A], DC [auth D] | PHEOPHYTIN A C55 H74 N4 O5 CQIKWXUXPNUNDV-RCBXBCQGSA-N |  | ||
| SQD Query on SQD | DA [auth A], SC [auth M] | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL C41 H78 O12 S RVUUQPKXGDTQPG-JUDHQOGESA-N |  | ||
| LMG Query on LMG | AC [auth C] BC [auth C] CB [auth B] EA [auth A] GB [auth C] | 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE C45 H86 O10 DCLTVZLYPPIIID-CVELTQQQSA-N |  | ||
| PL9 Query on PL9 | HC [auth D] | 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE C53 H80 O2 FKUYMLZIRPABFK-UHFFFAOYSA-N |  | ||
| LHG Query on LHG | HA [auth A], IC [auth D], JC [auth D], LC [auth D], RC [auth L] | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE C38 H75 O10 P BIABMEZBCHDPBV-MPQUPPDSSA-N |  | ||
| 3PH Query on 3PH | EB [auth B] | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE C39 H77 O8 P YFWHNAWEOZTIPI-DIPNUNPCSA-N |  | ||
| DGA Query on DGA | FB [auth B] | DIACYL GLYCEROL C39 H76 O5 UHUSDOQQWJGJQS-QNGWXLTQSA-N |  | ||
| HEM Query on HEM | MC [auth E] | PROTOPORPHYRIN IX CONTAINING FE C34 H32 Fe N4 O4 KABFMIBPWCXCRK-RGGAHWMASA-L |  | ||
| C7Z Query on C7Z | BB [auth B] | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol C40 H56 O2 JKQXZKUSFCKOGQ-ANDPMPNWSA-N |  | ||
| RRX Query on RRX | NC [auth H] | (3R)-beta,beta-caroten-3-ol C40 H56 O DMASLKHVQRHNES-FKKUPVFPSA-N |  | ||
| BCR Query on BCR | AB [auth B] CA [auth A] GC [auth D] PC [auth J] UB [auth C] | BETA-CAROTENE C40 H56 OENHQHLEOONYIE-JLTXGRSLSA-N |  | ||
| LMU Query on LMU | QC [auth J] | DODECYL-ALPHA-D-MALTOSIDE C24 H46 O11 NLEBIOOXCVAHBD-YHBSTRCHSA-N |  | ||
| OEX Query on OEX | U [auth A] | CA-MN4-O5 CLUSTER Ca Mn4 O5 SEXWDHMBWJEXOJ-UHFFFAOYSA-N |  | ||
| BCT Query on BCT | GA [auth A] | BICARBONATE ION C H O3 BVKZGUZCCUSVTD-UHFFFAOYSA-M |  | ||
| FE2 Query on FE2 | V [auth A] | FE (II) ION Fe CWYNVVGOOAEACU-UHFFFAOYSA-N |  | ||
| CL Query on CL | W [auth A], X [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| NA Query on NA | FA [auth A] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| CSD Query on CSD | B | L-PEPTIDE LINKING | C3 H7 N O4 S |  | CYS |
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | RELION | 4.0.0 |
Currently 9GNW does not have a validation slider image.
| Funding Organization | Location | Grant Number |
|---|---|---|
| Israel Science Foundation | Israel | 199/21 |