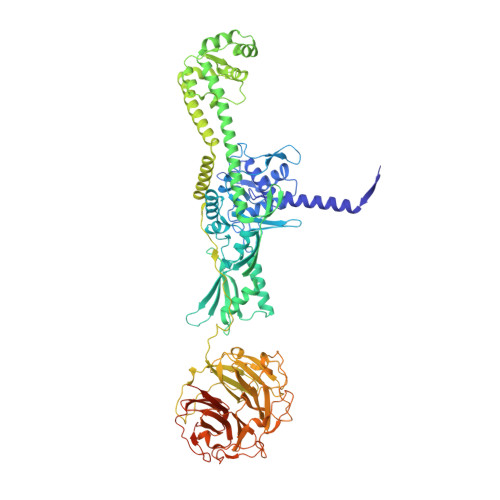
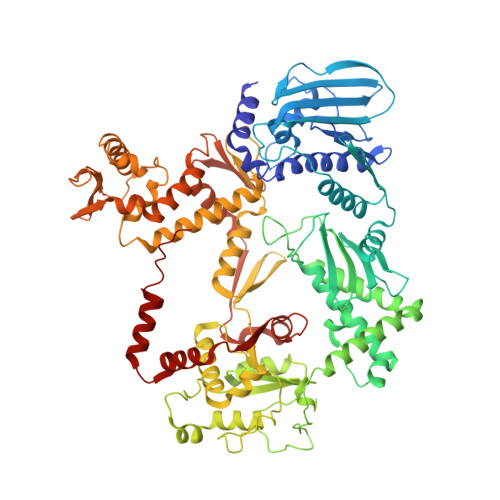
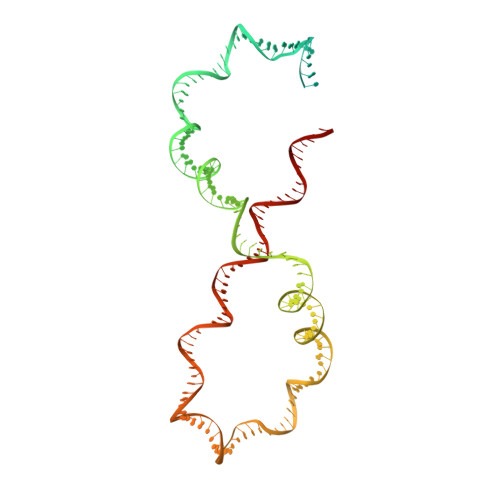
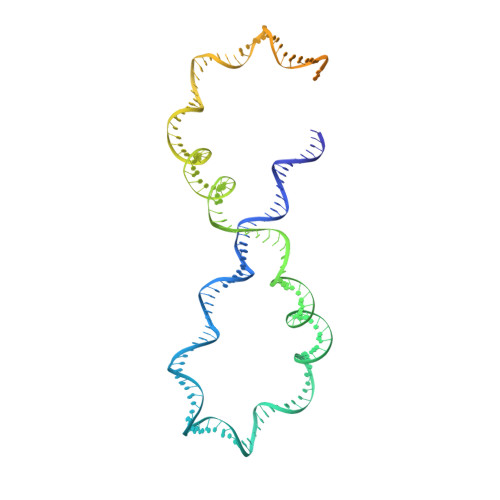
Structure of Escherichia coli DNA gyrase with chirally wrapped DNA supports ratchet-and-pawl mechanism for an ATP-powered supercoiling motor
Michalczyk, E., Pakosz-Stepien, Z., Liston, J., Gittins, O., Pabis, M., Heddle, J.G., Ghilarov, D.(2024) Proceedings Of The National Academy Of Sciences Usa