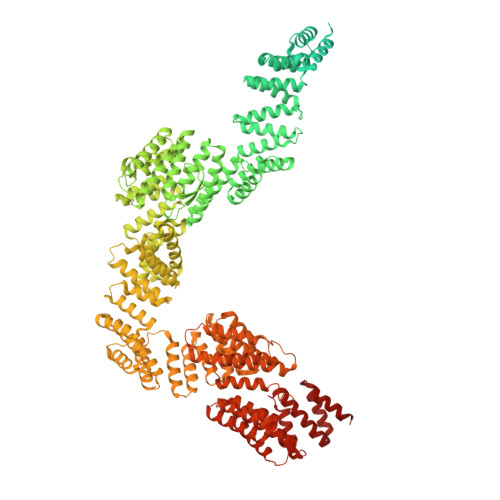
Structural basis of mRNA decay by the human exosome-ribosome supercomplex.
Kogel, A., Keidel, A., Loukeri, M.J., Kuhn, C.C., Langer, L.M., Schafer, I.B., Conti, E.(2024) Nature 635: 237-242
- PubMed: 39385025
- DOI: https://doi.org/10.1038/s41586-024-08015-6
- Primary Citation of Related Structures:
9G8M, 9G8N, 9G8O, 9G8P, 9G8Q, 9G8R - PubMed Abstract:
The interplay between translation and mRNA decay is widespread in human cells 1-3 . In quality-control pathways, exonucleolytic degradation of mRNA associated with translating ribosomes is mediated largely by the cytoplasmic exosome 4-9 , which includes the exoribonuclease complex EXO10 and the helicase complex SKI238 (refs. 10-16 ). The helicase can extract mRNA from the ribosome and is expected to transfer it to the exoribonuclease core through a bridging factor, HBS1L3 (also known as SKI7), but the mechanisms of this molecular handover remain unclear 7,17,18 . Here we reveal how human EXO10 is recruited by HBS1L3 (SKI7) to an active ribosome-bound SKI238 complex. We show that rather than a sequential handover, a direct physical coupling mechanism takes place, which culminates in the formation of a cytoplasmic exosome-ribosome supercomplex. Capturing the structure during active decay reveals a continuous path in which an RNA substrate threads from the 80S ribosome through the SKI2 helicase into the exoribonuclease active site of the cytoplasmic exosome complex. The SKI3 subunit of the complex directly binds to HBS1L3 (SKI7) and also engages a surface of the 40S subunit, establishing a recognition platform in collided disomes. Exosome and ribosome thus work together as a single structural and functional unit in co-translational mRNA decay, coordinating their activities in a transient supercomplex.
- Department of Structural Cell Biology, Max Planck Institute of Biochemistry, Martinsried, Germany.
Organizational Affiliation: