Nanobody-mediated activation and inhibition of Staphylococcus aureus MazF
Prolic-Kalinsek, M., Zorzini, V., Haesaerts, S., De Bruyn, P., Loris, R.To be published.
Experimental Data Snapshot
Starting Models: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Endoribonuclease MazF | 133 | Staphylococcus aureus | Mutation(s): 0 Gene Names: mazF, SA1873 EC: 3.1 | 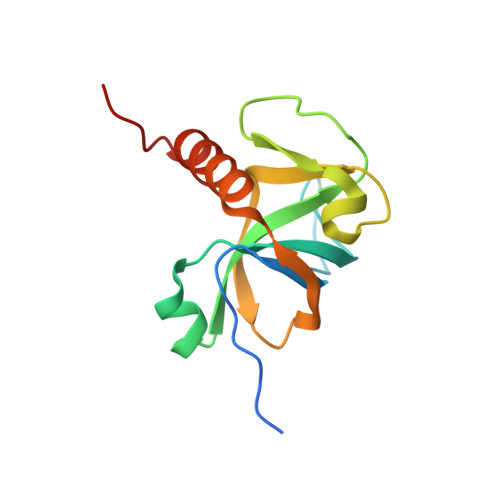 | |
UniProt | |||||
Find proteins for Q2FWI8 (Staphylococcus aureus (strain NCTC 8325 / PS 47)) Explore Q2FWI8 Go to UniProtKB: Q2FWI8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q2FWI8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Nanobody 12 | 136 | Lama glama | Mutation(s): 0 | 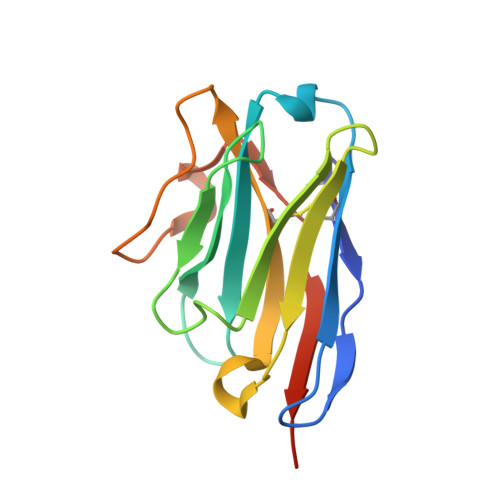 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| SO4 Query on SO4 | FA [auth G] GA [auth G] HA [auth G] IA [auth H] J [auth A] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| CL Query on CL | AA [auth G] BA [auth G] CA [auth G] DA [auth G] EA [auth G] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 52.19 | α = 89.85 |
| b = 61.27 | β = 83.89 |
| c = 97.31 | γ = 67.19 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| XDS | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Vrije Universiteit Brussel | Belgium | SPR13 |