New rhinovirus uncoating intermediate reveals how sodium versus potassium ions influence RNA release.
Real-Hohn, A., Blaas, D.(2025) Sci Rep 15: 36768-36768
- PubMed: 41120457
- DOI: https://doi.org/10.1038/s41598-025-20627-0
- Primary Citation of Related Structures:
9G0B - PubMed Abstract:
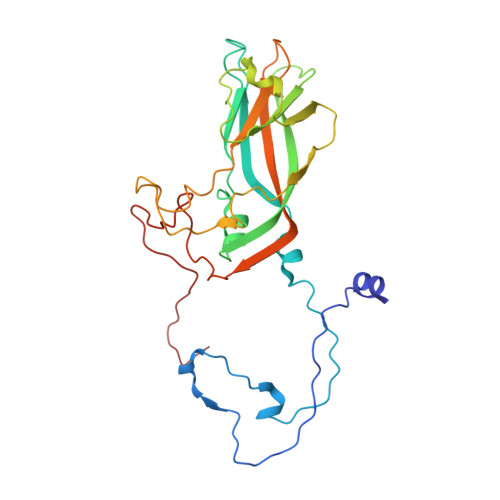
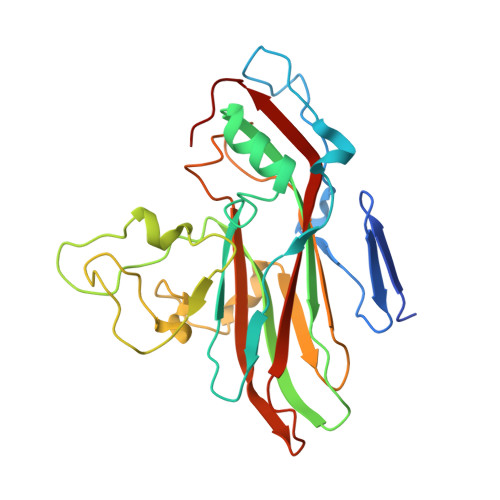
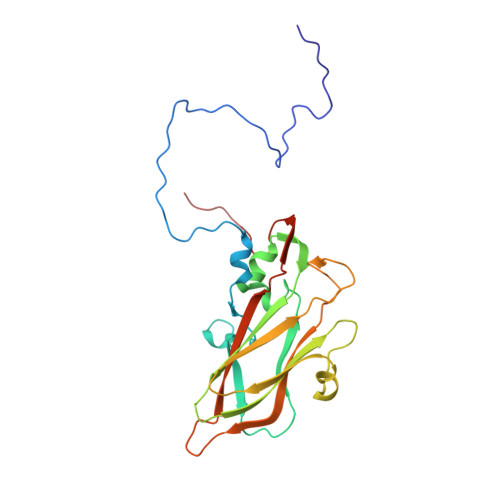
Electron microscopy (EM) of rhinovirus A2 (RV-A2) incubated in Na + phosphate buffer (pH 7.6) for 12 h at 25 °C revealed partial fragmentation, whereas upon incubation in K + phosphate buffer, RV-A2 appeared intact. In buffers adjusted to pH 5.8, these differences became more pronounced; acidic Na + phosphate buffer promoted disintegration of the particles, whereas in acidic K + phosphate buffer, the virus appeared like native. Incubation in the acidic buffers for one hour at 4 °C followed by neutralisation resulted in the respective formation of non-infectious A particles (in Na + ) and a non-infectious novel uncoating intermediate (in K + ), which we termed 'E0 particle'. Negative staining EM revealed phosphotungstate penetration into A particles, but not into E0 particles. Cryo-EM image reconstruction of the E0 particle showed clear differences between A and E0 particles; like native virus, E0 contained VP4 and a pocket factor. Native RV-A2 RNA cores, obtained by gentle proteinase-K digestion in K + and Na + phosphate buffer, respectively, differed in accessibility of dsRNA regions, detected by PaSTRy. Variance in RNA compactness observed in K + versus Na + phosphate buffer was confirmed by rotary shadowing EM; in K + phosphate buffer, the RNA remained condensed while, in Na + phosphate buffer, distinct unfolding stages were apparent.
- Centre for Medical Biochemistry, Max Perutz Labs, Vienna BioCenter, Medical University of Vienna, Vienna, Austria. antonio@realhohnlabs.com.
Organizational Affiliation: