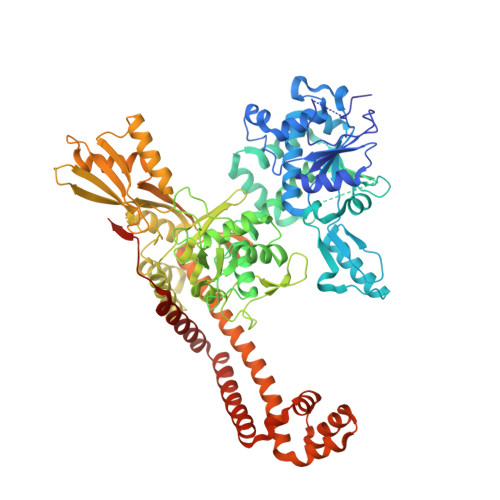
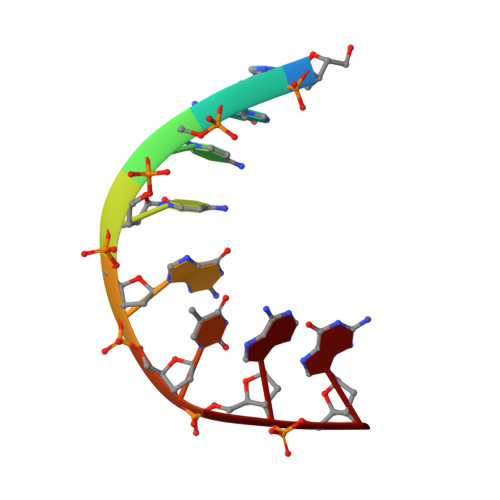
Structural Aspects of Mycobacterium tuberculosis DNA Gyrase Targeted by Novel Bacterial Topoisomerase Inhibitors.
Kokot, M., Hrast Rambaher, M., Feng, L., Mitchenall, L.A., Lawson, D.M., Maxwell, A., Parish, T., Minovski, N., Anderluh, M.(2024) ACS Med Chem Lett 15: 2164-2170
- PubMed: 39691510
- DOI: https://doi.org/10.1021/acsmedchemlett.4c00447
- Primary Citation of Related Structures:
9FOY - PubMed Abstract:
In this Letter, we present a small series of novel bacterial topoisomerase inhibitors (NTBIs) that exhibit both potent inhibition of Mycobacterium tuberculosis DNA gyrase and potent antimycobacterial activity. The disclosed crystal structure of M. tuberculosis DNA gyrase in complex with DNA and compound 5 from this NBTI series reveals the binding mode of an NBTI in the GyrA binding pocket and confirms the presence and importance of halogen bonding for the excellent on-target potency. In addition, we have shown that compound 5 is a promising M. tuberculosis DNA gyrase inhibitor, with an IC 50 for M. tuberculosis gyrase of 0.096 μM, and it has potent activity against M. tuberculosis , with an IC 50 of 0.165 μM.
- Theory Department, Laboratory for Cheminformatics, National Institute of Chemistry, Hajdrihova 19, 1001 Ljubljana, Slovenia.
Organizational Affiliation: