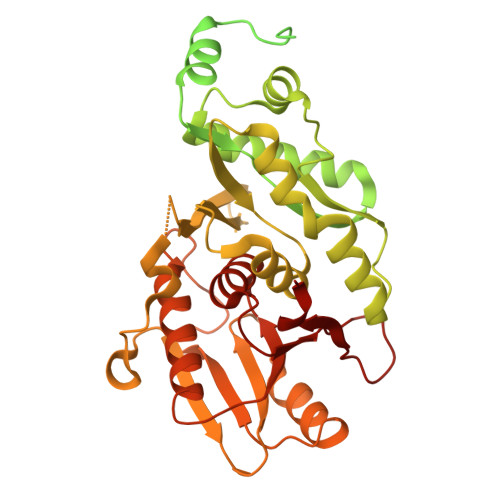
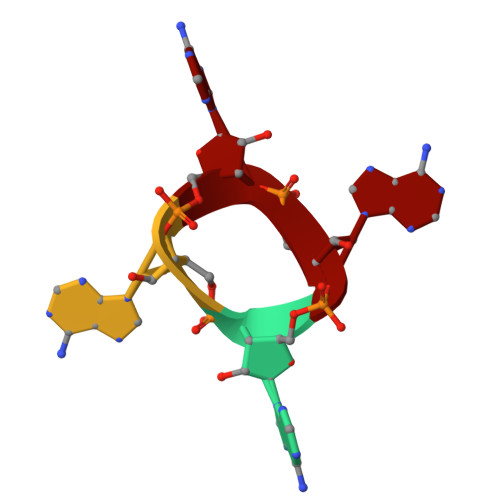
Filament formation activates protease and ring nuclease activities of CRISPR Lon-SAVED.
Smalakyte, D., Ruksenaite, A., Sasnauskas, G., Tamulaitiene, G., Tamulaitis, G.(2024) Mol Cell 84: 4239-4255.e8
- PubMed: 39362215
- DOI: https://doi.org/10.1016/j.molcel.2024.09.002
- Primary Citation of Related Structures:
9EYI, 9EYJ, 9EYK - PubMed Abstract:
To combat phage infection, type III CRISPR-Cas systems utilize cyclic oligoadenylates (cA n ) signaling to activate various auxiliary effectors, including the CRISPR-associated Lon-SAVED protease CalpL, which forms a tripartite effector system together with an anti-σ factor, CalpT, and an ECF-like σ factor, CalpS. Here, we report the characterization of the Candidatus Cloacimonas acidaminovorans CalpL-CalpT-CalpS. We demonstrate that cA 4 binding triggers CalpL filament formation and activates it to cleave CalpT within the CalpT-CalpS dimer. This cleavage exposes the CalpT C-degron, which targets it for further degradation by cellular proteases. Consequently, CalpS is released to bind to RNA polymerase, causing growth arrest in E. coli. Furthermore, the CalpL-CalpT-CalpS system is regulated by the SAVED domain of CalpL, which is a ring nuclease that cleaves cA 4 in a sequential three-step mechanism. These findings provide key mechanistic details for the activation, proteolytic events, and regulation of the signaling cascade in the type III CRISPR-Cas immunity.
- Institute of Biotechnology, Life Sciences Center, Vilnius University, Saulėtekio av. 7, 10257 Vilnius, Lithuania.
Organizational Affiliation: