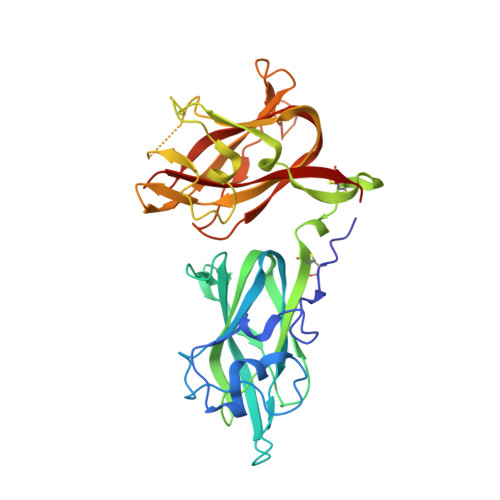
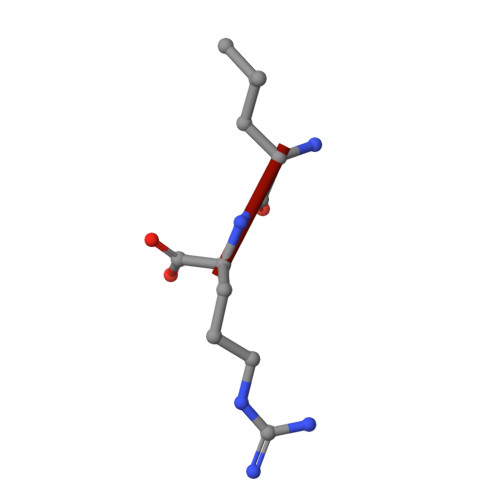
Identification of an osteopontin-derived peptide that binds neuropilin-1 and activates vascular repair responses and angiogenesis.
Chen, Y., Gialeli, C., Shen, J., Duner, P., Walse, B., Duelli, A., Caing-Carlsson, R., Blom, A.M., Zibert, J.R., Nilsson, A.H., Alenfall, J., Liang, C., Nilsson, J.(2024) Pharmacol Res 205: 107259-107259
- PubMed: 38871237
- DOI: https://doi.org/10.1016/j.phrs.2024.107259
- Primary Citation of Related Structures:
9EOU - PubMed Abstract:
The osteopontin-derived peptide FOL-005 stimulates hair growth. Using ligand-receptor glyco-capture technology we identified neuropilin-1 (NRP-1), a known co-receptor for vascular endothelial growth factor (VEGF) receptors, as the most probable receptor for FOL-005 and the more stable analogue FOL-026. X-ray diffraction and microscale thermophoresis analysis revealed that FOL-026 shares binding site with VEGF in the NRP-1 b1-subdomain. Stimulation of human umbilical vein endothelial cells with FOL-026 resulted in phosphorylation of VEGFR-2, ERK1/2 and AKT, increased cell growth and migration, stimulation of endothelial tube formation and inhibition of apoptosis in vitro. FOL-026 also promoted angiogenesis in vivo as assessed by subcutaneous Matrigel plug and hind limb ischemia models. NRP-1 knock-down or treatment of NRP-1 antagonist EG00229 blocked the stimulatory effects of FOL-026 on endothelial cells. Exposure of human coronary artery smooth muscle cells to FOL-026 stimulated cell growth, migration, inhibited apoptosis, and induced VEGF gene expression and VEGFR-2/AKT phosphorylation by an NRP-1-dependent mechanism. RNA sequencing showed that FOL-026 activated pathways involved in tissue repair. These findings identify NRP-1 as the receptor for FOL-026 and show that its biological effects mimic that of growth factors binding to the VEGF receptor family. They also suggest that FOL-026 may have therapeutical potential in conditions that require vascular repair and/or enhanced angiogenesis.
- Department of Cardiology, Second Affiliated Hospital of Naval Medical University, Shanghai Cardiovascular Institute of Integrative Medicine, Shanghai, China; Department of Experimental Medical Science, Lund University, Sweden.
Organizational Affiliation: