Structural basis of RNA-mediated regulation of transcriptional pausing.
Porta, J.C., Ellinger, E., Liu, Y., Chauvier, A., Walter, N.G.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA-directed RNA polymerase subunit alpha | C [auth G], D [auth H] | 231 | Escherichia coli | Mutation(s): 0 Gene Names: rpoA, Z4665, ECs4160 EC: 2.7.7.6 | 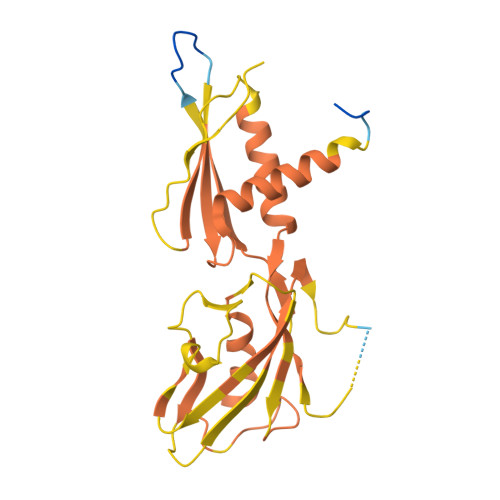 |
UniProt | |||||
Find proteins for P0A7Z4 (Escherichia coli (strain K12)) Explore P0A7Z4 Go to UniProtKB: P0A7Z4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A7Z4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA-directed RNA polymerase subunit beta | E [auth I] | 1,340 | Escherichia coli | Mutation(s): 0 Gene Names: rpoB EC: 2.7.7.6 | 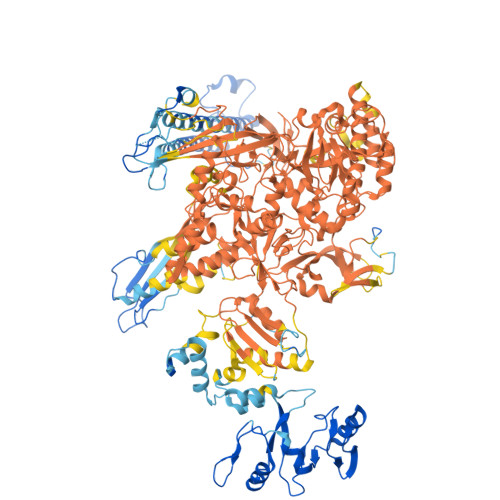 |
UniProt | |||||
Find proteins for P0A8V2 (Escherichia coli (strain K12)) Explore P0A8V2 Go to UniProtKB: P0A8V2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A8V2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA-directed RNA polymerase subunit beta' | F [auth J] | 1,358 | Escherichia coli | Mutation(s): 0 Gene Names: rpoC EC: 2.7.7.6 | 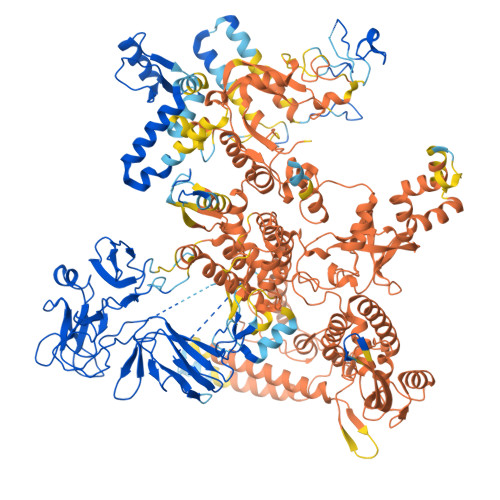 |
UniProt | |||||
Find proteins for P0A8T7 (Escherichia coli (strain K12)) Explore P0A8T7 Go to UniProtKB: P0A8T7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A8T7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA-directed RNA polymerase subunit omega | G [auth K] | 79 | Escherichia coli | Mutation(s): 0 Gene Names: rpoZ, Ecok1_36260, APECO1_2812 EC: 2.7.7.6 | 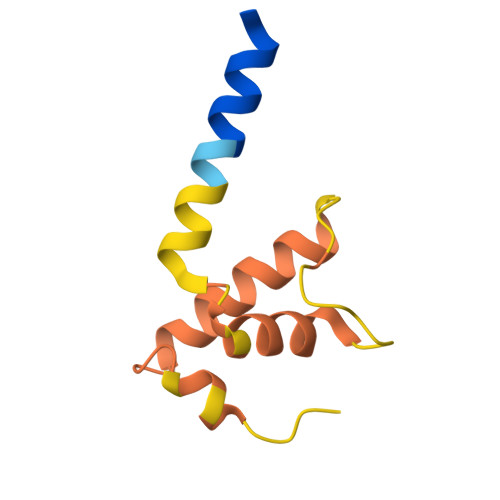 |
UniProt | |||||
Find proteins for P0A800 (Escherichia coli (strain K12)) Explore P0A800 Go to UniProtKB: P0A800 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A800 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (39-MER) | 30 | Escherichia coli |  | ||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (30-MER) | 30 | Escherichia coli |  | ||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| RNA (5'-R(P*UP*GP*GP*UP*AP*GP*GP*AP*GP*U)-3') | H [auth R] | 10 | Escherichia coli |  | |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| MG Query on MG | I [auth J] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.20_4459: |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | R35 GM131922 |