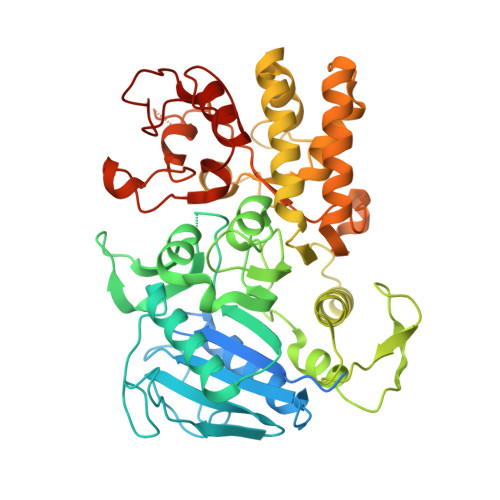
MicroED structure of the C11 cysteine protease clostripain.
Ruma, Y.N., Bu, G., Hattne, J., Gonen, T.(2024) J Struct Biol X 10: 100107-100107
- PubMed: 39100863
- DOI: https://doi.org/10.1016/j.yjsbx.2024.100107
- Primary Citation of Related Structures:
9CIP - PubMed Abstract:
Clostripain secreted from Clostridium histolyticum is the founding member of the C11 family of Clan CD cysteine peptidases, which is an important group of peptidases secreted by numerous bacteria. Clostripain is an arginine-specific endopeptidase. Because of its efficacy as a cysteine peptidase, it is widely used in laboratory settings. Despite its importance the structure of clostripain remains unsolved. Here we describe the first structure of an active form of C. histolyticum clostripain determined at 2.5 Å resolution using microcrystal electron diffraction (MicroED). The structure was determined from a single nanocrystal after focused ion beam milling. The structure of clostripain shows a typical Clan CD α/β/α sandwich architecture and the Cys231/His176 catalytic dyad in the active site. It has a large electronegative substrate binding pocket showing its ability to accommodate large and diverse substrates. A loop in the heavy chain formed between residues 452 and 457 is potentially important for substrate binding. In conclusion, this result demonstrates the importance of MicroED to determine the unknown structure of macromolecules such as clostripain, which can be further used as a platform to study substrate binding and design of potential inhibitors against this class of peptidases.
- Howard Hughes Medical Institute, University of California, Los Angeles, Los Angeles, CA 90095, United States.
Organizational Affiliation: