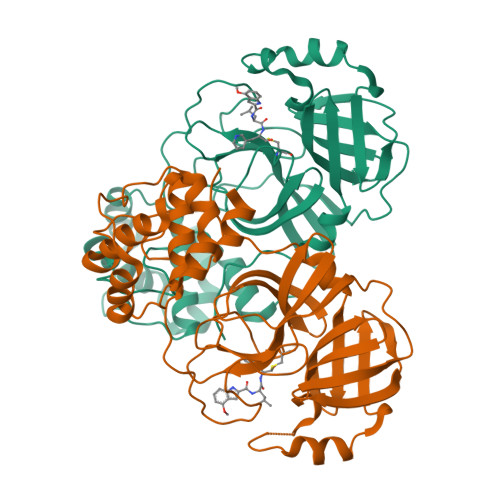
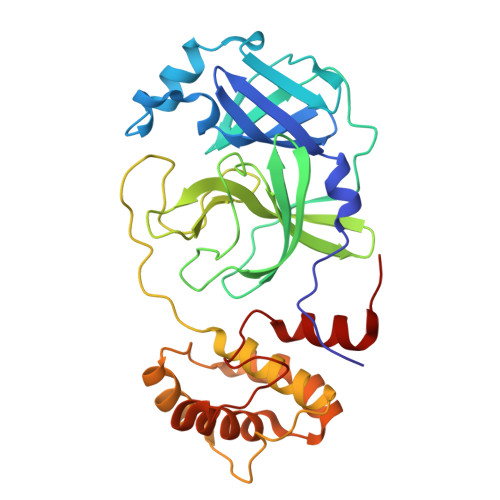
Dual Inhibitors of SARS-CoV-2 3CL Protease and Human Cathepsin L Containing Glutamine Isosteres Are Anti-CoV-2 Agents.
Kumar, V., Zhu, J., Chenna, B.C., Hoffpauir, Z.A., Rademacher, A., Rogers, A.M., Tseng, C.T., Drelich, A., Farzandh, S., Lamb, A.L., Meek, T.D.(2025) J Am Chem Soc 147: 1631-1648
- PubMed: 39746101
- DOI: https://doi.org/10.1021/jacs.4c11620
- Primary Citation of Related Structures:
9CEC, 9CED, 9CEK, 9CF9, 9CFB - PubMed Abstract:
SARS-CoV-2 3CL protease (Main protease) and human cathepsin L are proteases that play unique roles in the infection of human cells by SARS-CoV-2, the causative agent of COVID-19. Both proteases recognize leucine and other hydrophobic amino acids at the P 2 position of a peptidomimetic inhibitor. At the P 1 position, cathepsin L accepts many amino acid side chains, with a partial preference for phenylalanine, while 3CL-PR protease has a stringent specificity for glutamine or glutamine analogues. We have designed, synthesized, and evaluated peptidomimetic aldehyde dual-target (dual-acting) inhibitors using two peptide scaffolds based on those of two Pfizer 3CL-PR inhibitors, Nirmatrelvir , and PF-835321 . Our inhibitors contain glutamine isosteres at the P 1 position, including 2-pyridon-3-yl-alanine, 3-pyridinyl-alanine, and 1,3-oxazo-4-yl-alanine groups. Inhibition constants for these new inhibitors ranged from K i = 0.6-18 nM (cathepsin L) and K i = 2.6-124 nM (3CL-PR), for which inhibitors with the 2-pyridon-3-yl-alanal substituent were the most potent for 3CL-PR. The anti-CoV-2 activity of these inhibitors ranged from EC 50 = 0.47-15 μM. X-ray structures of the peptidomimetic aldehyde inhibitors of 3CL-PR with similar scaffolds all demonstrated the formation of thiohemiacetals with Cys 145 , and hydrogen-bonding interactions with the heteroatoms of the pyridon-3-yl-alanyl group, as well as the nitrogen of the N-terminal indole and its appended carbonyl group at the P 3 position. The absence of these hydrogen bonds for the inhibitors containing the 3-pyridinyl-alanyl and 1,3-oxazo-4-yl-alanyl groups was reflected in the less potent inhibition of the inhibitors with 3CL-PR. In summary, our studies demonstrate the value of a second generation of cysteine protease inhibitors that comprise a single agent that acts on both human cathepsin L and SARS-CoV-2 3CL protease. Such dual-target inhibitors will provide anti-COVID-19 drugs that remain active despite the development of resistance due to mutation of the viral protease. Such dual-target inhibitors are more likely to remain useful therapeutics despite the emergence of inactivating mutations in the viral protease because the human cathepsin L will not develop resistance. This particular dual-target approach is innovative since one of the targets is viral (3CL-PR) required for viral protein maturation and the other is human (hCatL) which enables viral infection.
Organizational Affiliation:
Department of Biochemistry and Biophysics, Texas A&M University, 301 Old Main Drive, College Station, Texas 77845, United States.