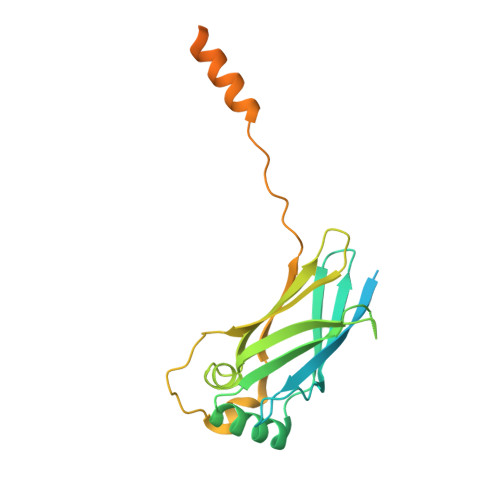
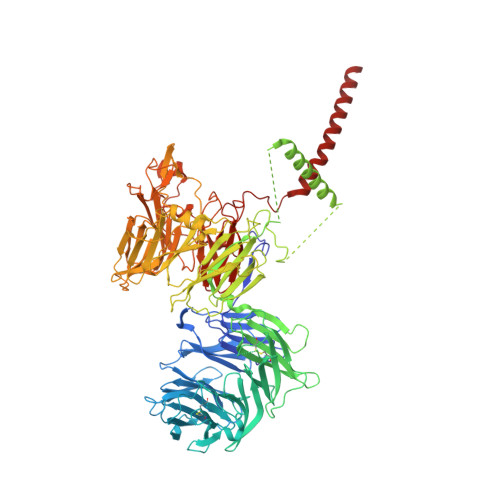
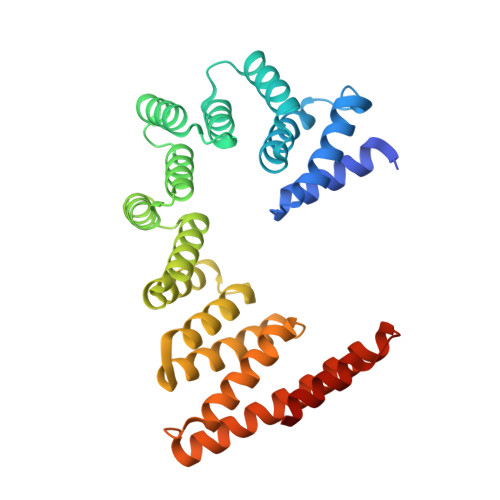
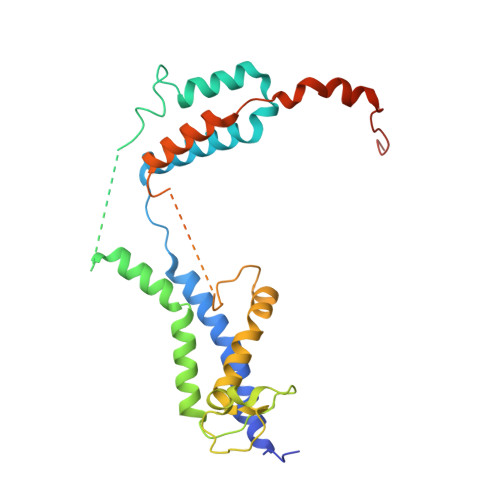
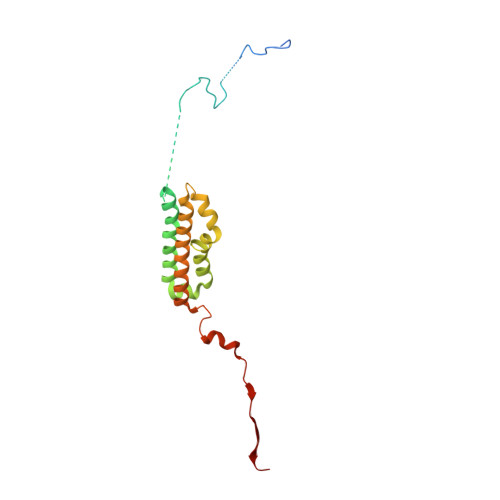
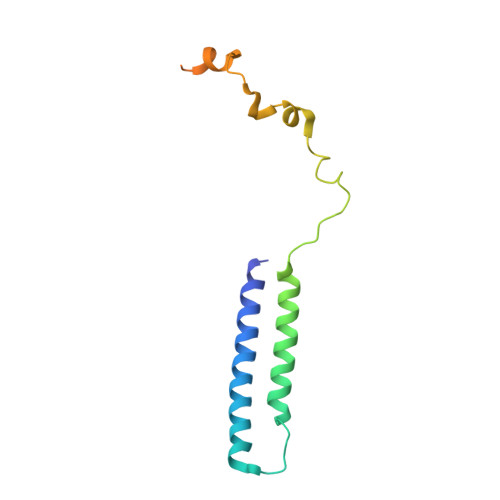
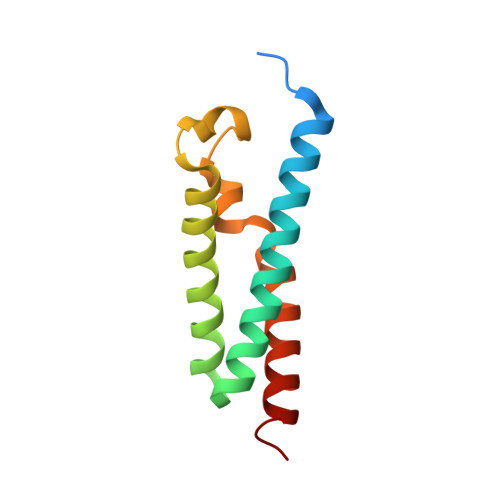
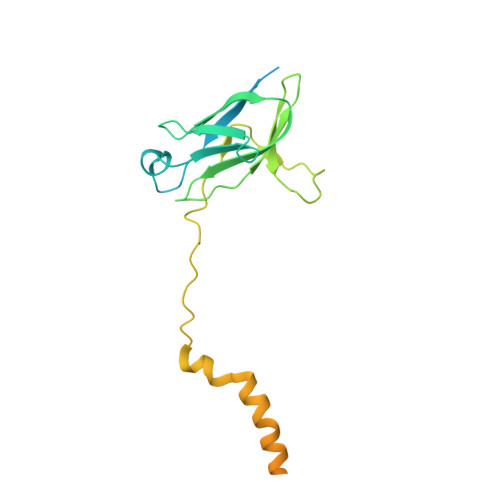
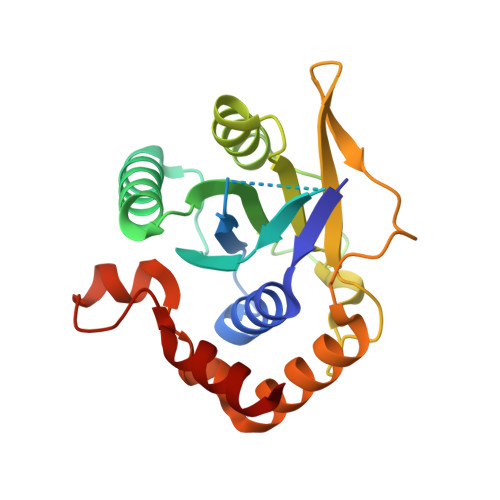
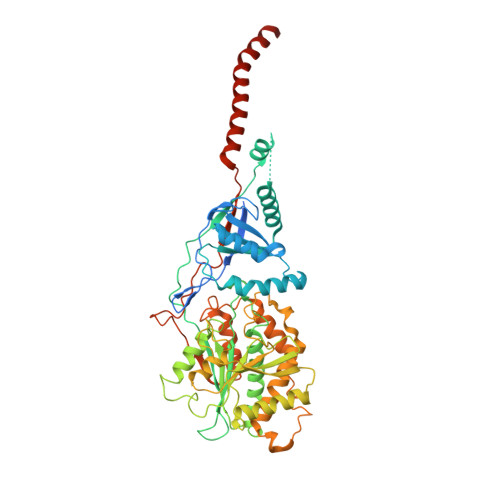
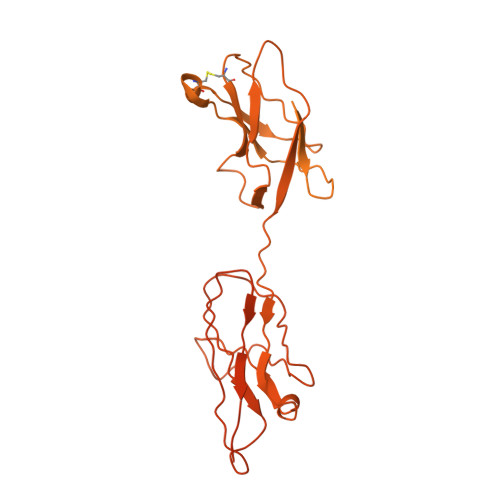
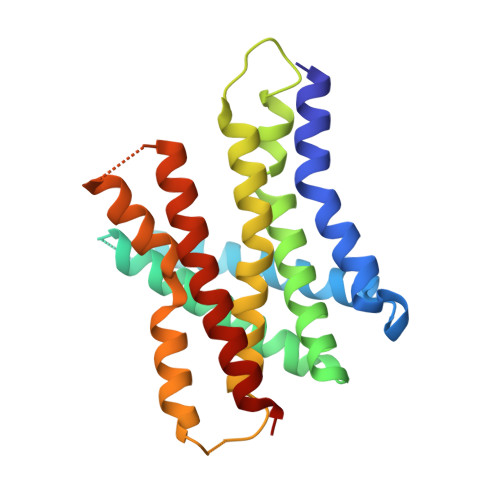
Role of a holo-insertase complex in the biogenesis of biophysically diverse ER membrane proteins.
Page, K.R., Nguyen, V.N., Pleiner, T., Tomaleri, G.P., Wang, M.L., Guna, A., Hazu, M., Wang, T.Y., Chou, T.F., Voorhees, R.M.(2024) Mol Cell 84: 3302
- PubMed: 39173640
- DOI: https://doi.org/10.1016/j.molcel.2024.08.005
- Primary Citation of Related Structures:
9C7U, 9C7V - PubMed Abstract:
Mammalian membrane proteins perform essential physiologic functions that rely on their accurate insertion and folding at the endoplasmic reticulum (ER). Using forward and arrayed genetic screens, we systematically studied the biogenesis of a panel of membrane proteins, including several G-protein-coupled receptors (GPCRs). We observed a central role for the insertase, the ER membrane protein complex (EMC), and developed a dual-guide approach to identify genetic modifiers of the EMC. We found that the back of Sec61 (BOS) complex, a component of the multipass translocon, was a physical and genetic interactor of the EMC. Functional and structural analysis of the EMC⋅BOS holocomplex showed that characteristics of a GPCR's soluble domain determine its biogenesis pathway. In contrast to prevailing models, no single insertase handles all substrates. We instead propose a unifying model for coordination between the EMC, the multipass translocon, and Sec61 for the biogenesis of diverse membrane proteins in human cells.
- Division of Biology and Biological Engineering, California Institute of Technology, 1200 E. California Ave., Pasadena, CA 91125, USA.
Organizational Affiliation: