Structure and function of XMAP215s C terminal domains in microtubule nucleation
McManus, C.T., Travis, S.M., Jeffrey, P.D., Zhang, R., Petry, S.To be published.
Experimental Data Snapshot
Starting Models: in silico, experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Tubulin beta-2B chain | A [auth B], B [auth D] | 445 | Bos taurus | Mutation(s): 0 | 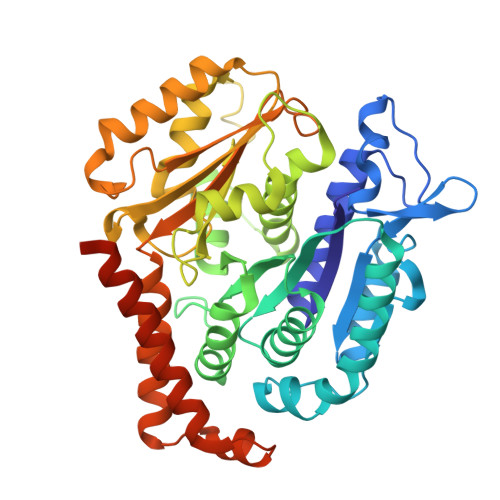 |
UniProt | |||||
Find proteins for Q6B856 (Bos taurus) Explore Q6B856 Go to UniProtKB: Q6B856 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q6B856 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Tubulin alpha chain | C [auth A], D [auth C] | 451 | Bos taurus | Mutation(s): 0 | 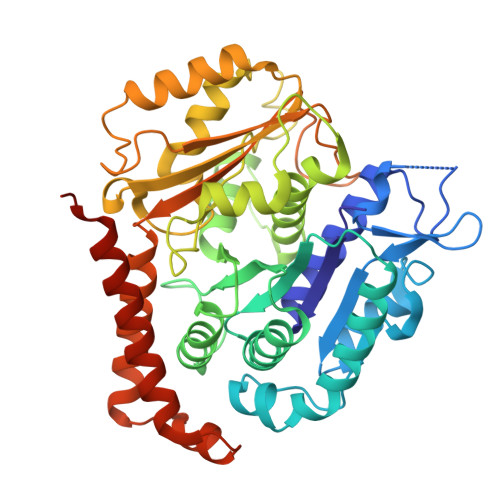 |
UniProt | |||||
Find proteins for F2Z4C1 (Bos taurus) Explore F2Z4C1 Go to UniProtKB: F2Z4C1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | F2Z4C1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytoskeleton-associated protein 5-A,Cytoskeleton-associated protein 5-A,Green fluorescent protein | 541 | Xenopus laevis | Mutation(s): 0 Gene Names: ckap5-a, xmap215, GFP | 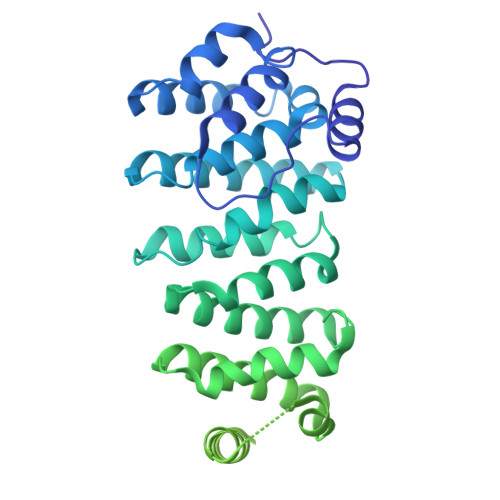 | |
UniProt | |||||
Find proteins for Q9PT63 (Xenopus laevis) Explore Q9PT63 Go to UniProtKB: Q9PT63 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9PT63 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| GTP Query on GTP | J [auth A], L [auth C] | GUANOSINE-5'-TRIPHOSPHATE C10 H16 N5 O14 P3 XKMLYUALXHKNFT-UUOKFMHZSA-N |  | ||
| G2P Query on G2P | F [auth B], H [auth D] | PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER C11 H18 N5 O13 P3 GXTIEXDFEKYVGY-KQYNXXCUSA-N |  | ||
| MG Query on MG | G [auth B], I [auth D], K [auth A], M [auth C] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.21-5207-000 |
| RECONSTRUCTION | cryoSPARC | 3.3.2 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | 1R01 GM141100-01A1 |
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | F32 GM142149-01A1 |
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | 1R01 GM141100-01A1 |