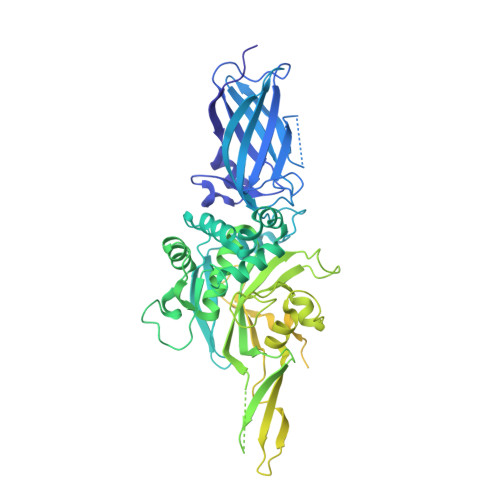
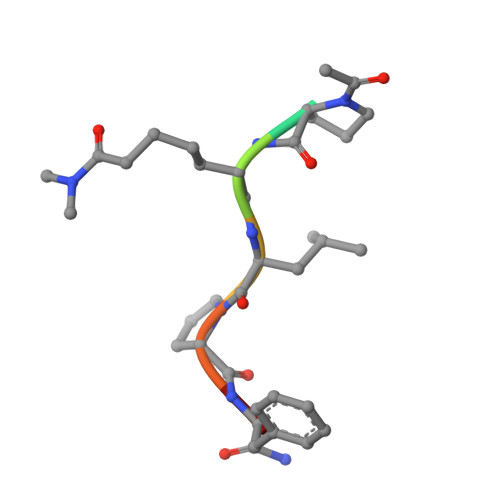
Structural and mechanistic analysis of Ca 2+ -dependent regulation of transglutaminase 2 activity using a Ca 2+ -bound intermediate state.
Sewa, A.S., Besser, H.A., Mathews, I.I., Khosla, C.(2024) Proc Natl Acad Sci U S A 121: e2407066121-e2407066121
- PubMed: 38959038
- DOI: https://doi.org/10.1073/pnas.2407066121
- Primary Citation of Related Structures:
9BC2, 9BC3, 9BC4 - PubMed Abstract:
Mammalian transglutaminases, a family of Ca 2+ -dependent proteins, are implicated in a variety of diseases. For example, celiac disease (CeD) is an autoimmune disorder whose pathogenesis requires transglutaminase 2 (TG2) to deamidate select glutamine residues in diet-derived gluten peptides. Deamidation involves the formation of transient γ-glutamyl thioester intermediates. Recent studies have revealed that in addition to the deamidated gluten peptides themselves, their corresponding thioester intermediates are also pathogenically relevant. A mechanistic understanding of this relevance is hindered by the absence of any structure of Ca 2+ -bound TG2. We report the X-ray crystallographic structure of human TG2 bound to an inhibitory gluten peptidomimetic and two Ca 2+ ions in sites previously designated as S1 and S3. Together with additional structure-guided experiments, this structure provides a mechanistic explanation for how S1 regulates formation of an inhibitory disulfide bond in TG2, while also establishing that S3 is essential for γ-glutamyl thioester formation. Furthermore, our crystallographic findings and associated analyses have revealed that i) two interacting residues, H305 and E363, play a critical role in resolving the thioester intermediate into an isopeptide bond (transamidation) but not in thioester hydrolysis (deamidation); and ii) residues N333 and K176 stabilize preferred TG2 substrates and inhibitors via hydrogen bonding to nonreactive backbone atoms. Overall, the intermediate-state conformer of TG2 reported here represents a superior model to previously characterized conformers for both transition states of the TG2-catalyzed reaction.
Organizational Affiliation:
Department of Biochemistry, Stanford University School of Medicine, Stanford, CA 94305.