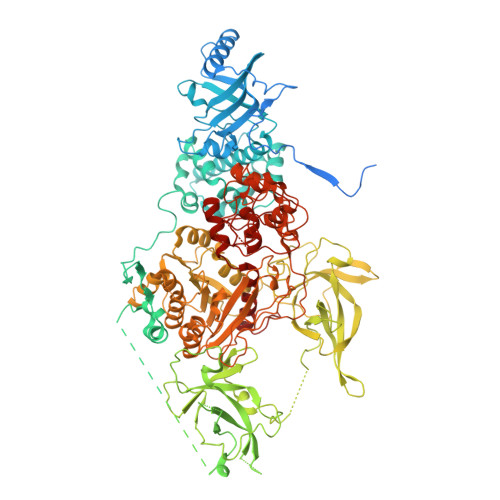
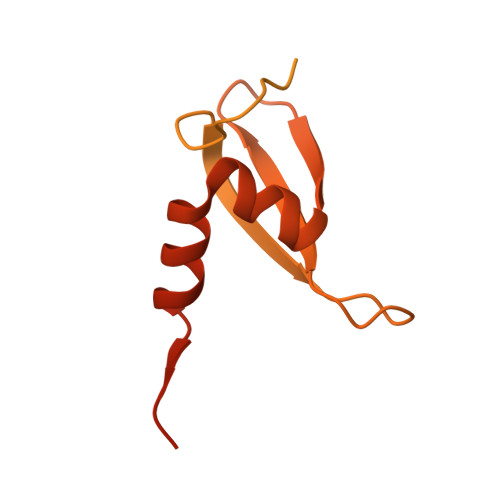
Multi-layered heterochromatin interaction as a switch for DIM2-mediated DNA methylation.
Shao, Z., Lu, J., Khudaverdyan, N., Song, J.(2024) Nat Commun 15: 6815-6815
- PubMed: 39122718
- DOI: https://doi.org/10.1038/s41467-024-51246-4
- Primary Citation of Related Structures:
9BAP, 9BAQ, 9BAZ - PubMed Abstract:
Functional crosstalk between DNA methylation, histone H3 lysine-9 trimethylation (H3K9me3) and heterochromatin protein 1 (HP1) is essential for proper heterochromatin assembly and genome stability. However, how repressive chromatin cues guide DNA methyltransferases for region-specific DNA methylation remains largely unknown. Here, we report structure-function characterizations of DNA methyltransferase Defective-In-Methylation-2 (DIM2) in Neurospora. The DNA methylation activity of DIM2 requires the presence of both H3K9me3 and HP1. Our structural study reveals a bipartite DIM2-HP1 interaction, leading to a disorder-to-order transition of the DIM2 target-recognition domain that is essential for substrate binding. Furthermore, the structure of DIM2-HP1-H3K9me3-DNA complex reveals a substrate-binding mechanism distinct from that for its mammalian orthologue DNMT1. In addition, the dual recognition of H3K9me3 peptide by the DIM2 RFTS and BAH1 domains allosterically impacts the DIM2-substrate binding, thereby controlling DIM2-mediated DNA methylation. Together, this study uncovers how multiple heterochromatin factors coordinately orchestrate an activity-switching mechanism for region-specific DNA methylation.
- Department of Biochemistry, University of California, Riverside, CA, 92521, USA.
Organizational Affiliation: