CryoEM structure of DIM2-HP1-H3K9me3-DNA complex
Song, J., Shao, Z.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA (cytosine-5-)-methyltransferase | 1,244 | Neurospora crassa | Mutation(s): 0 Gene Names: dim-2, GE21DRAFT_10473 EC: 2.1.1.37 | 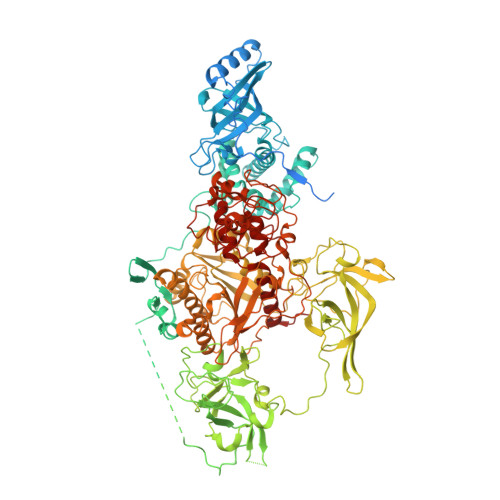 | |
UniProt | |||||
Find proteins for Q96W73 (Neurospora crassa) Explore Q96W73 Go to UniProtKB: Q96W73 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q96W73 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Heterochromatin protein one | 268 | Neurospora crassa | Mutation(s): 0 Gene Names: 49D12.150, hpo, GE21DRAFT_9232 | 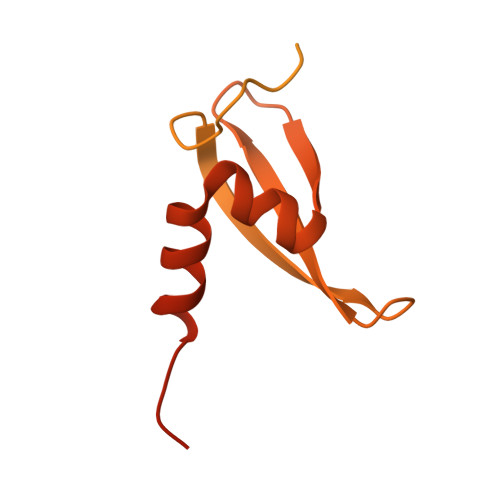 | |
UniProt | |||||
Find proteins for Q870N8 (Neurospora crassa) Explore Q870N8 Go to UniProtKB: Q870N8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q870N8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Histone H3.2 | D, E [auth F] | 25 | Neurospora crassa | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q5MYA4 (Cichorium intybus) Explore Q5MYA4 Go to UniProtKB: Q5MYA4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5MYA4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(*AP*GP*TP*AP*GP*GP*AP*GP*GP*AP*GP*GP*AP*GP*TP*AP*GP*T)-3') | F [auth G] | 18 | Neurospora crassa |  | |
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(*AP*CP*TP*AP*CP*T)-R(P*(PYO))-D(P*CP*TP*CP*CP*TP*CP*CP*TP*AP*CP*T)-3') | G [auth H] | 18 | Neurospora crassa |  | |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| SAH Query on SAH | H [auth A] | S-ADENOSYL-L-HOMOCYSTEINE C14 H20 N6 O5 S ZJUKTBDSGOFHSH-WFMPWKQPSA-N |  | ||
| ZN Query on ZN | I [auth A] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| M3L Query on M3L | D, E [auth F] | L-PEPTIDE LINKING | C9 H21 N2 O2 |  | LYS |
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.21_5207: |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | -- |