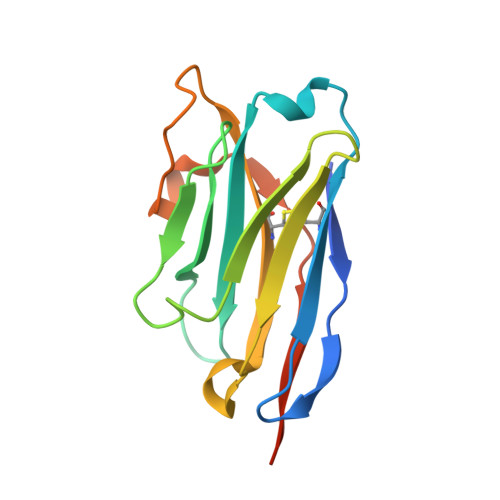
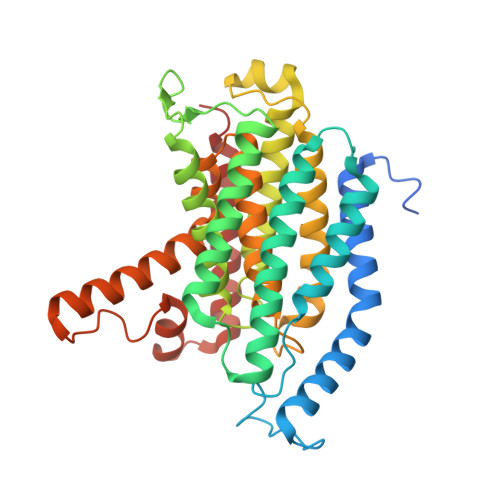
Development of a natural product optimization strategy for inhibitors against MraY, a promising antibacterial target.
Yamamoto, K., Sato, T., Hao, A., Asao, K., Kaguchi, R., Kusaka, S., Ruddarraju, R.R., Kazamori, D., Seo, K., Takahashi, S., Horiuchi, M., Yokota, S.I., Lee, S.Y., Ichikawa, S.(2024) Nat Commun 15: 5085-5085
- PubMed: 38877016
- DOI: https://doi.org/10.1038/s41467-024-49484-7
- Primary Citation of Related Structures:
9B70, 9B71 - PubMed Abstract:
MraY (phospho-N-acetylmuramoyl-pentapeptide-transferase) inhibitory natural products are attractive molecules as candidates for a new class of antibacterial agents to combat antimicrobial-resistant bacteria. Structural optimization of these natural products is required to improve their drug-like properties for therapeutic use. However, chemical modifications of these natural products are painstaking tasks due to complex synthetic processes, which is a bottleneck in advancing natural products to the clinic. Here, we develop a strategy for a comprehensive in situ evaluation of the build-up library, which enables us to streamline the preparation of the analogue library and directly assess its biological activities. We apply this approach to a series of MraY inhibitory natural products. Through construction and evaluation of the 686-compound library, we identify promising analogues that exhibit potent and broad-spectrum antibacterial activity against highly drug-resistant strains in vitro as well as in vivo in an acute thigh infection model. Structures of the MraY-analogue complexes reveal distinct interaction patterns, suggesting that these analogues represent MraY inhibitors with unique binding modes. We further demonstrate the generality of our strategy by applying it to tubulin-binding natural products to modulate their tubulin polymerization activities.
- Faculty of Pharmaceutical Sciences, Hokkaido University, Kita-12, Nishi-6, Kita-ku, Sapporo, 060-0812, Japan. k.yamamoto@pharm.hokudai.ac.jp.
Organizational Affiliation: