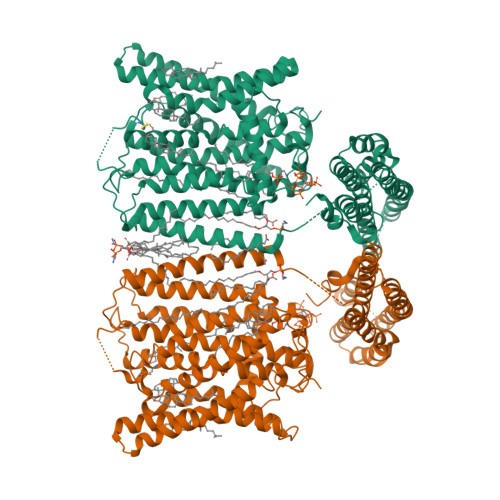
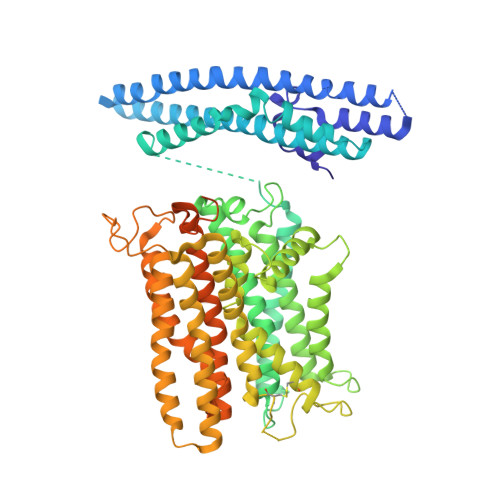
Synergistic activation of the human phosphate exporter XPR1 by KIDINS220 and inositol pyrophosphate.
Zuo, P., Wang, W., Dai, Z., Zheng, J., Yu, S., Wang, G., Yin, Y., Liang, L., Yin, Y.(2025) Nat Commun 16: 2879-2879
- PubMed: 40128258
- DOI: https://doi.org/10.1038/s41467-025-58200-y
- Primary Citation of Related Structures:
9INE, 9INF, 9INH, 9ITG, 9IUC - PubMed Abstract:
Inorganic phosphate (Pi) is essential for life, and its intracellular levels must be tightly regulated to avoid toxicity. XPR1, the sole known phosphate exporter, is critical for maintaining this balance. Here we report cryo-EM structures of the human XPR1-KIDINS220 complex in substrate-free closed and substrate-bound outward-open states, as well as an XPR1 mutant in a substrate-bound inward-facing state. In the presence of inositol hexaphosphate (InsP 6 ) and phosphate, the complex adopts an outward-open conformation, with InsP 6 binding the SPX domain and juxtamembrane regions, indicating active phosphate export. Without phosphate or InsP 6 , the complex closes, with transmembrane helix 9 blocking the outward cavity and a C-terminal loop obstructing the intracellular cavity. XPR1 alone remains closed even with phosphate and InsP 6 . Functional mutagenesis shows that InsP 6 , whose levels vary with Pi availability, works with KIDINS220 to regulate XPR1 activity. These insights into phosphate regulation may aid in developing therapies for ovarian cancer.
Organizational Affiliation:
Institute of Systems Biomedicine, Department of Pathology, Beijing Key Laboratory of Tumor Systems Biology, School of Basic Medical Sciences, Peking University Health Science Center, Beijing, China.