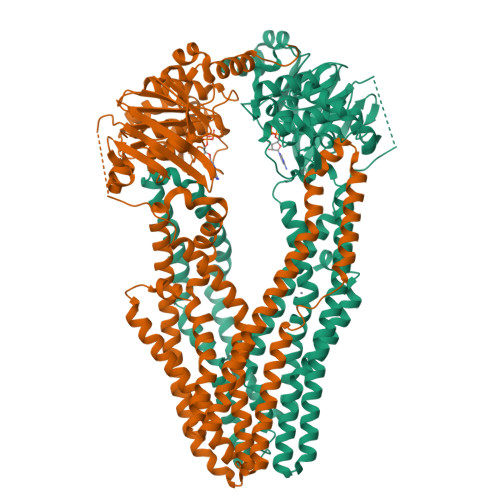
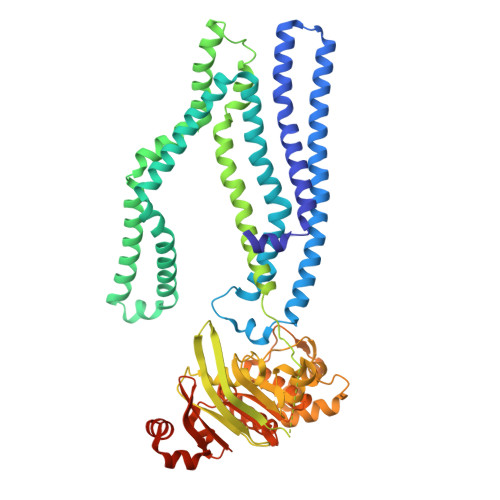
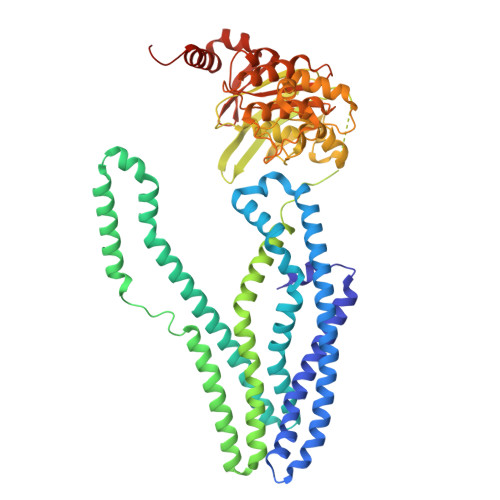
The mycobacterial ABC transporter IrtAB employs a membrane-facing crevice for siderophore-mediated iron uptake.
Gonda, I., Sorrentino, S., Galazzo, L., Lichti, N.P., Arnold, F.M., Mehdipour, A.R., Bordignon, E., Seeger, M.A.(2025) Nat Commun 16: 1133-1133
- PubMed: 39880813
- DOI: https://doi.org/10.1038/s41467-024-55136-7
- Primary Citation of Related Structures:
9FW3, 9FXC, 9G2K, 9G2L, 9G2M, 9G2P, 9G2S, 9G2T, 9G2V, 9G2X, 9G2Y, 9G2Z, 9G36, 9G37, 9GL3 - PubMed Abstract:
The mycobacterial ABC transporter IrtAB features an ABC exporter fold, yet it imports iron-charged siderophores called mycobactins. Here, we present extensive cryo-EM analyses and DEER measurements, revealing that IrtAB alternates between an inward-facing and an outward-occluded conformation, but does not sample an outward-facing conformation. When IrtAB is locked in its outward-occluded conformation in nanodiscs, mycobactin is bound in the middle of the lipid bilayer at a membrane-facing crevice opening at the heterodimeric interface. Mutations introduced at the crevice abrogate mycobactin import and in corresponding structures, the crevice is collapsed. A conserved triple histidine motif coordinating a zinc ion is present below the mycobactin binding site. Substitution of these histidine residues with alanine results in a decoupled transporter, which hydrolyzes ATP, but lost its capacity to import mycobactins. Our data suggest that IrtAB imports mycobactin via a credit-card mechanism in a transport cycle that is coupled to the presence of zinc.
Organizational Affiliation:
Institute of Medical Microbiology, University of Zurich, Zurich, Switzerland.