Structure-Based Optimization of a Series of Covalent, Cell Active Bfl-1 Inhibitors.
Lucas, S.C.C., Blackwell, J.H., Borjesson, U., Hargreaves, D., Milbradt, A.G., Bostock, M.J., Ahmed, S., Beaumont, K., Cheung, T., Demanze, S., Gohlke, A., Guerot, C., Haider, A., Kantae, V., Kauffman, G.W., Kinzel, O., Kupcova, L., Lainchbury, M.D., Lamb, M.L., Leon, L., Palisse, A., Sacchetto, C., Storer, R.I., Su, N., Thomson, C., Vales, J., Chen, Y., Hu, X.(2024) J Med Chem 67: 16455-16479
- PubMed: 39291659
- DOI: https://doi.org/10.1021/acs.jmedchem.4c01288
- Primary Citation of Related Structures:
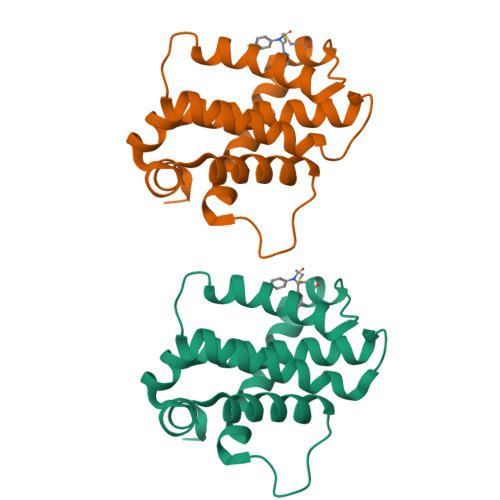
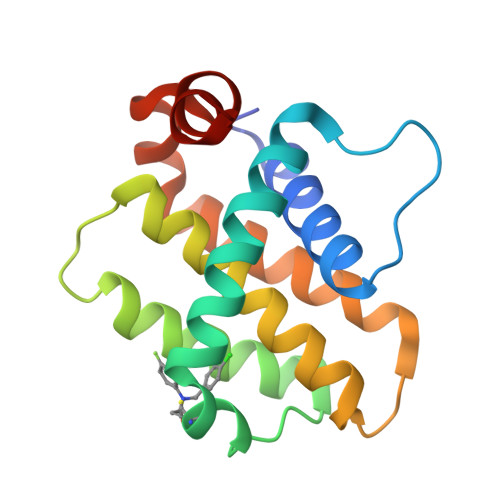
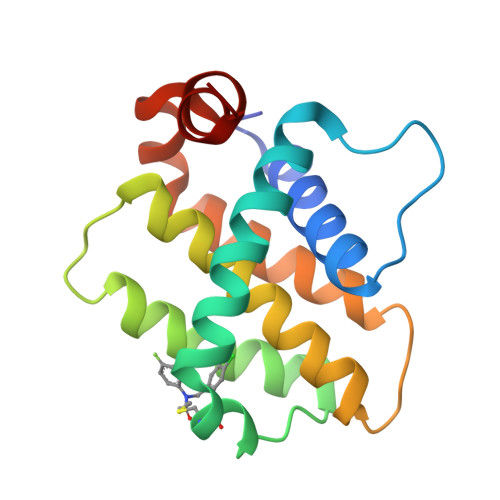
9FKY, 9FKZ, 9FL0 - PubMed Abstract:
Bfl-1, a member of the Bcl-2 family of proteins, plays a crucial role in apoptosis regulation and has been implicated in cancer cell survival and resistance to venetoclax therapy. Due to the unique cysteine residue in the BH3 binding site, the development of covalent inhibitors targeting Bfl-1 represents a promising strategy for cancer treatment. Herein, the optimization of a covalent cellular tool from a lead-like hit using structure based design is described. Informed by a reversible X-ray fragment screen, the strategy to establish interactions with a key glutamic acid residue (Glu78) and optimize binding in a cryptic pocket led to a 1000-fold improvement in biochemical potency without increasing reactivity of the warhead. Compound ( R,R,S )-26 has a k inact /K I of 4600 M -1 s -1 , shows <1 μM caspase activation in a cellular assay and cellular target engagement, and has good physicochemical properties and a promising in vivo profile.
Organizational Affiliation:
Hit Discovery, Discovery Sciences, R&D, AstraZeneca, Cambridge CB2 0AA, U.K.