Structural dynamics of DNA unwinding by a replicative helicase.
Shahid, T., Danazumi, A.U., Tehseen, M., Alhudhali, L., Clark, A.R., Savva, C.G., Hamdan, S.M., De Biasio, A.(2025) Nature
- PubMed: 40108462
- DOI: https://doi.org/10.1038/s41586-025-08766-w
- Primary Citation of Related Structures:
9EVH, 9EVP, 9EXD, 9F3T, 9F3U, 9F5I, 9F73, 9F74, 9F75, 9F7N, 9F9N, 9F9O, 9F9W, 9F9X, 9FA1, 9FA2, 9FB0, 9FB4, 9FB5, 9FB6, 9KAE, 9KAK - PubMed Abstract:
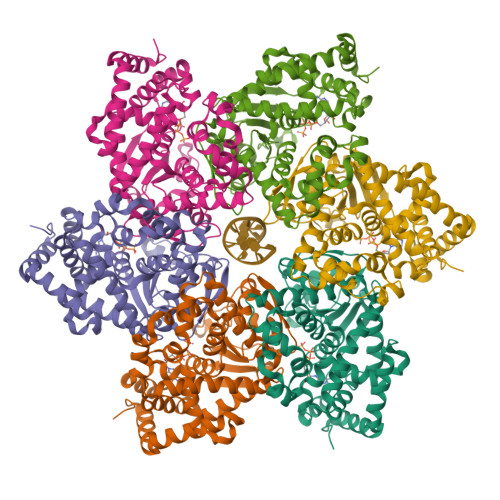
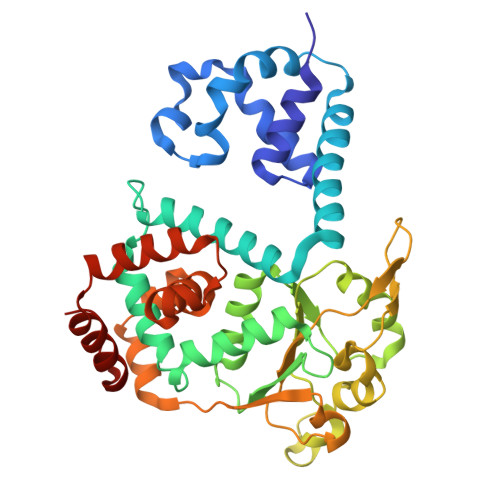
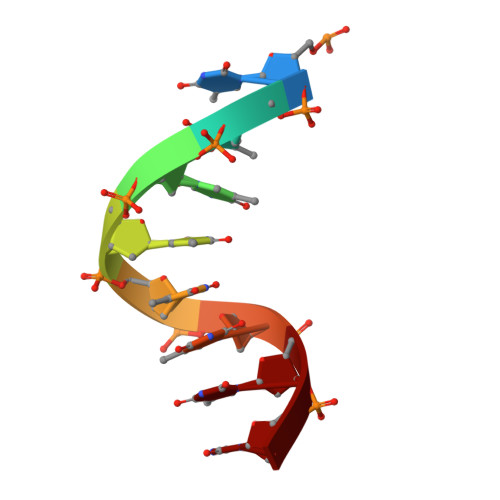
Hexameric helicases are nucleotide-driven molecular machines that unwind DNA to initiate replication across all domains of life. Despite decades of intensive study, several critical aspects of their function remain unresolved 1 : the site and mechanism of DNA strand separation, the mechanics of unwinding propagation, and the dynamic relationship between nucleotide hydrolysis and DNA movement. Here, using cryo-electron microscopy (cryo-EM), we show that the simian virus 40 large tumour antigen (LTag) helicase assembles in the form of head-to-head hexamers at replication origins, melting DNA at two symmetrically positioned sites to establish bidirectional replication forks. Through continuous heterogeneity analysis 2 , we characterize the conformational landscape of LTag on forked DNA under catalytic conditions, demonstrating coordinated motions that drive DNA translocation and unwinding. We show that the helicase pulls the tracking strand through DNA-binding loops lining the central channel, while directing the non-tracking strand out of the rear, in a cyclic process. ATP hydrolysis functions as an 'entropy switch', removing blocks to translocation rather than directly powering DNA movement. Our structures show the allosteric couplings between nucleotide turnover and subunit motions that enable DNA unwinding while maintaining dedicated exit paths for the separated strands. These findings provide a comprehensive model for replication fork establishment and progression that extends from viral to eukaryotic systems. More broadly, they introduce fundamental principles of the mechanism by which ATP-dependent enzymes achieve efficient mechanical work through entropy-driven allostery.
Organizational Affiliation:
Bioscience Program, Division of Biological and Environmental Sciences and Engineering, King Abdullah University of Science and Technology, Thuwal, Saudi Arabia.