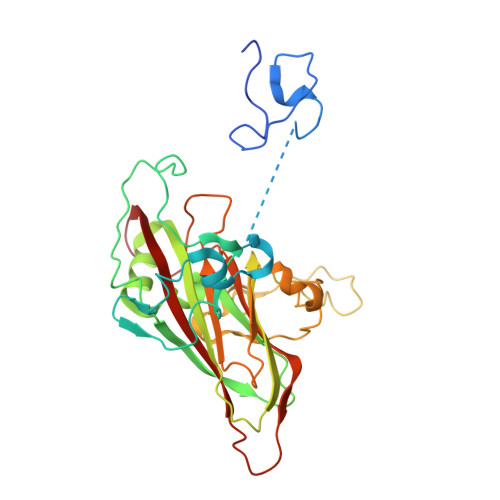
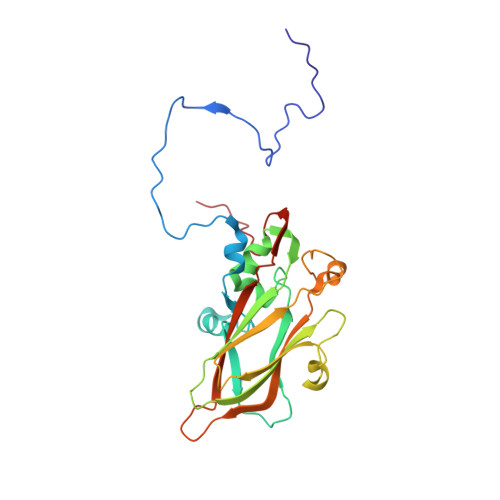
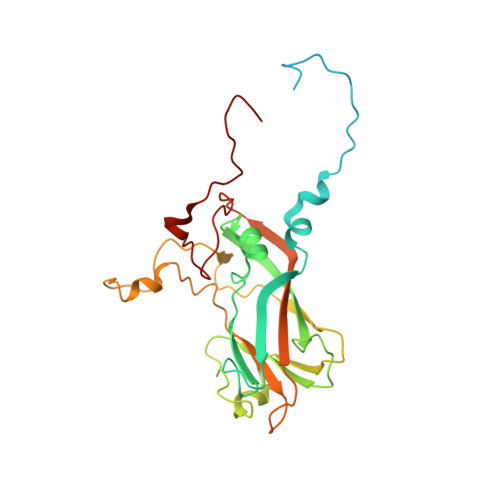
Reappraisal of the mechanism of the enterovirus VP0 maturation cleavage based on the structure of a stabilised assembly intermediate
Kingston, N.J., Snowden, J.S.S., Grehan, K.G., Hall, P.K., Hietanen, E.V., Passchier, T.C., Polyak, S.J., Filman, D.J., Hogle, J.M., Rowlands, D.J., Stonehouse, N.J.To be published.