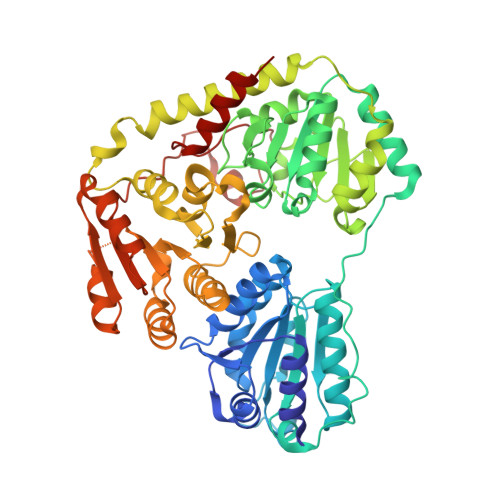
Corynebacterium glutamicum pyruvate:quinone oxidoreductase: an enigmatic metabolic enzyme with unusual structural features.
da Silva Lameira, C., Munssinger, S., Yang, L., Eikmanns, B.J., Bellinzoni, M.(2024) FEBS J
- PubMed: 39080980
- DOI: https://doi.org/10.1111/febs.17232
- Primary Citation of Related Structures:
9EV3, 9EV4, 9EV5, 9EV6 - PubMed Abstract:
Pyruvate:quinone oxidoreductase (PQO) is a flavin-containing peripheral membrane enzyme catalyzing the decarboxylation of pyruvate to acetate and CO 2 with quinone as an electron acceptor. Here, we investigate PQO activity in Corynebacterium glutamicum, examine purified PQO, and describe the crystal structure of the native enzyme and a truncated version. The specific PQO activity was highest in stationary phase cells grown in complex medium, lower in cells grown in complex medium containing glucose or acetate, and lowest in cells grown in minimal acetate-medium. A similar pattern with about 30-fold higher specific PQO activities was observed in C. glutamicum with plasmid-bound pqo expression under the control of the tac promoter, indicating that the differences in PQO activity are likely due to post-transcriptional control. Continuous cultivation of C. glutamicum at dilution rates between 0.05 and 0.4 h -1 revealed a negative correlation between PQO activity and growth rate. Kinetic analysis of PQO enzymes purified from cells grown in complex or in minimal acetate-medium revealed substantial differences in specific activity (72.3 vs. 11.9 U·mg protein -1 ) and turnover number (k cat : 440 vs. 78 s -1 , respectively), suggesting post-translational modifications affecting PQO activity. Structural analysis of PQO revealed a homotetrameric arrangement very similar to the Escherichia coli pyruvate oxidase PoxB except for the C-terminal membrane binding domain, which exhibited a conformation markedly different from its PoxB counterpart. A truncated PQO variant lacking 17 C-terminal amino acids showed higher affinity to pyruvate and was independent of detergent activation, highlighting the importance of the C-terminus for enzyme activation and lipid binding.
Organizational Affiliation:
Institute of Molecular Biology and Biotechnology of Prokaryotes, University of Ulm, Germany.