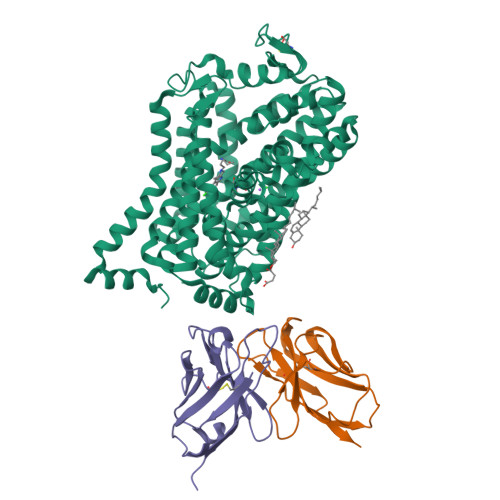
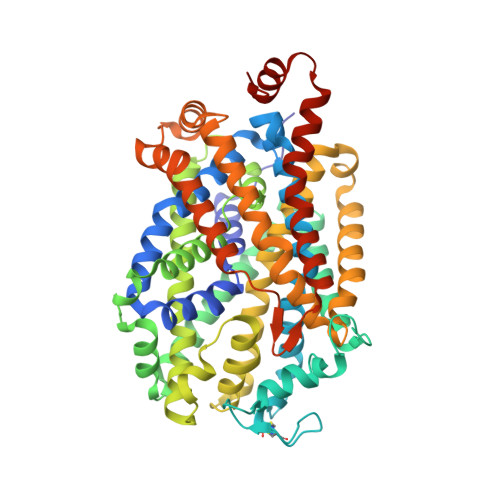
Cryo-EM structure of the dopamine transporter with a novel atypical non-competitive inhibitor bound to the orthosteric site.
Pedersen, C.N., Yang, F., Ita, S., Xu, Y., Akunuri, R., Trampari, S., Neumann, C.M.T., Desdorf, L.M., Schiott, B., Salvino, J.M., Mortensen, O.V., Nissen, P., Shahsavar, A.(2024) J Neurochem 168: 2043-2055
- PubMed: 39010681
- DOI: https://doi.org/10.1111/jnc.16179
- Primary Citation of Related Structures:
9EUO, 9EUP - PubMed Abstract:
The regulation of dopamine (DA) removal from the synaptic cleft is a crucial process in neurotransmission and is facilitated by the sodium- and chloride-coupled dopamine transporter DAT. Psychostimulant drugs, cocaine, and amphetamine, both block the uptake of DA, while amphetamine also triggers the release of DA. As a result, they prolong or even amplify neurotransmitter signaling. Atypical inhibitors of DAT lack cocaine-like rewarding effects and offer a promising strategy for the treatment of drug use disorders. Here, we present the 3.2 Å resolution cryo-electron microscopy structure of the Drosophila melanogaster dopamine transporter (dDAT) in complex with the atypical non-competitive inhibitor AC-4-248. The inhibitor partially binds at the central binding site, extending into the extracellular vestibule, and locks the transporter in an outward open conformation. Our findings propose mechanisms for the non-competitive inhibition of DAT and attenuation of cocaine potency by AC-4-248 and provide a basis for the rational design of more efficacious atypical inhibitors.
Organizational Affiliation:
DANDRITE, Nordic EMBL Partnership for Molecular Medicine, Department of Molecular Biology and Genetics, Aarhus University, Aarhus, Denmark.