Structural and functional insights into UDP-N-acetylglucosamine-enolpyruvate reductase (MurB) from Brucella ovis.
Minjarez-Saenz, M., Rivero, M., Correa-Perez, V., Boneta, S., Suarez, P., Polo, V., Sadeghi, S.J., Yruela, I., Martinez-Julvez, M., Medina, M.(2025) Arch Biochem Biophys 765: 110288-110288
- PubMed: 39761724
- DOI: https://doi.org/10.1016/j.abb.2025.110288
- Primary Citation of Related Structures:
9DTK - PubMed Abstract:
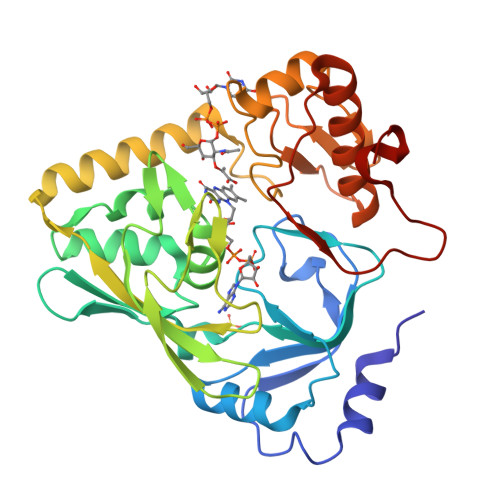
The peptidoglycan biosynthetic pathway involves a series of enzymatic reactions in which UDP-N-acetylglucosamine-enolpyruvate reductase (MurB) plays a crucial role in catalyzing the conversion of UDP-N-acetylglucosamine-enolpyruvate (UNAGEP) to UDP-N-acetylmuramic acid. This reaction relies on NADPH and FAD and, since MurB is not found in eukaryotes, it is an attractive target for the development of antimicrobials. MurB from Brucella ovis, the causative agent of brucellosis in sheep, is characterized here. The FAD cofactor in MurB of B. ovis is reduced to the hydroquinone state without semiquinone stabilization with an estimated E ox/hq of -260 mV. MurB from B. ovis catalyzes the oxidation of NADPH in a slow process that is positively influenced by the presence of the second product, UNAGEP. The crystallographic structure of the MurB ox :UNAGEP complex confirms its folding into three domains and the binding of UNAGEP, positioning its enolpyruvyl group for hydride transfer from FAD. MurB shows a complex thermal unfolding pathway that is influenced by UNAGEP and NADP + , confirming its ability to bind both molecules. Molecular dynamics (MD) simulations predict that the nicotinamide of NADP + is more stable at the active site than the enolpyruvyl of UNAGEP, and suggests that MurB can simultaneously accommodate NADPH and UNAGEP in the substrate channel, increasing overall protein-ligand flexibility. Sequence and evolutionary analyses show that MurB from B. ovis conserves all motifs predicted to be involved in catalysis within the Type IIa family.
Organizational Affiliation:
Departamento de Bioquímica y Biología Molecular y Celular, Facultad de Ciencias, Universidad de Zaragoza, Zaragoza, Spain; Instituto de Biocomputación y Física de Sistemas Complejos (BIFI) Universidad de Zaragoza, GBsC (Unizar) Join Unit to CSIC, Zaragoza, Spain.