Insights into structure and activity of a UDP-GlcNAc 2-epimerase involved in secondary cell wall polymer biosynthesis in Paenibacillus alvei.
Stefanovic, C., Legg, M.S.G., Mateyko, N., Ender, J.J., Kuvek, T., Oostenbrink, C., Schaffer, C., Evans, S.V., Hager-Mair, F.F.(2024) Front Mol Biosci 11: 1470989-1470989
- PubMed: 39391870
- DOI: https://doi.org/10.3389/fmolb.2024.1470989
- Primary Citation of Related Structures:
9CM8 - PubMed Abstract:
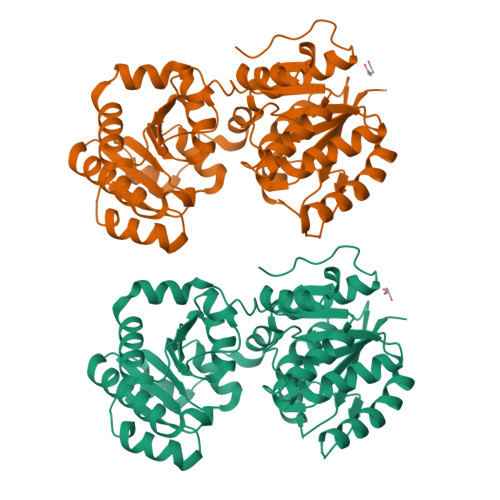
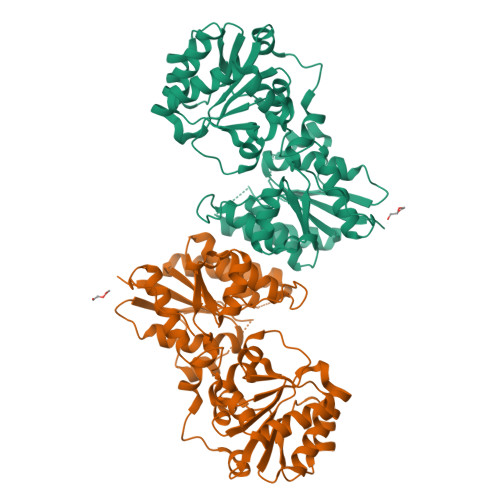
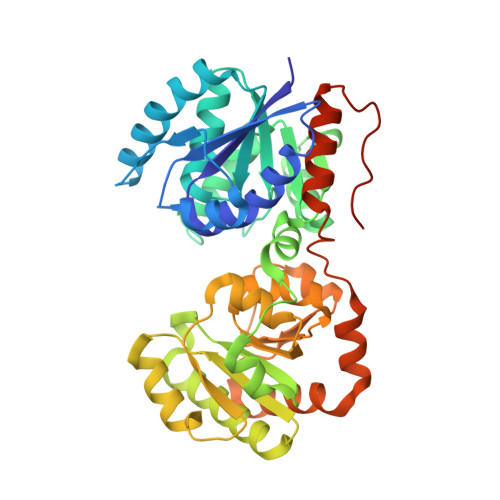
S-layer anchoring in Paenibacillus alvei is enabled by a non-covalent interaction between an S-layer homology domain trimer and a secondary cell wall polymer (SCWP), ensuring the structural integrity of the bacterial cell wall. Within the SCWP repeat, pyruvylated ManNAc serves as the ligand and the UDP-GlcNAc-2-epimerase MnaA supplies UDP-ManNAc to SCWP biosynthesis. To better understand SCWP biosynthesis and identify strategies for inhibiting pathogens with comparable cell wall architecture, like Bacillus anthracis , MnaA and rational variants were produced in E. coli and their kinetic constants determined. The effect of UDP-GlcNAc as a predicted allosteric activator and tunicamycin as a potential inhibitor of MnaA was tested in vitro supported by molecular docking experiments. Additionally, wild-type MnaA was crystallized. We present the crystal structure of unliganded P. alvei MnaA resolved at 2.20 Å. It adopts a GT-B fold consistent with other bacterial non-hydrolyzing UDP-GlcNAc 2-epimerases. A comparison of amino acid sequences reveals conservation of putative and known catalytic and allosteric-site residues in MnaA, which was confirmed through analysis of Q42A, Q69A, E135A and H241A MnaA variants. The kinetic parameters K M and k cat of MnaA were determined to be 3.91 mM and 33.44 s -1 for the forward, and 2.41 mM and 6.02 s -1 for the reverse reaction. While allosteric regulation by UDP-GlcNAc has been proposed as a mechanism for enzyme activation, UDP-GlcNAc was not found to be essential for UDP-ManNAc epimerization by P. alvei MnaA. However, the reaction rate doubled upon addition of 5% UDP-GlcNAc. Unexpectedly, the UDP-GlcNAc analog tunicamycin did not inhibit MnaA. Molecular docking experiments comparing tunicamycin binding of P. alvei MnaA and Staphylococcus aureus MnaA, which is inhibited by tunicamycin, revealed different residues exposed to the antibiotic excluding, those at the predicted allosteric site of P. alvei MnaA, corroborating tunicamycin resistance. The unliganded crystal structure of P. alvei MnaA reveals an open conformation characterized by an accessible cleft between the N- and C-terminal domains. Despite the conservation of residues involved in binding the allosteric activator UDP-GlcNAc, the enzyme is not strictly regulated by the substrate. Unlike S. aureus MnaA, the activity of P. alvei MnaA remains unaffected by tunicamycin.
- Department of Chemistry, Institute of Biochemistry, NanoGlycobiology Research Group, Universität für Bodenkultur Wien, Vienna, Austria.
Organizational Affiliation: