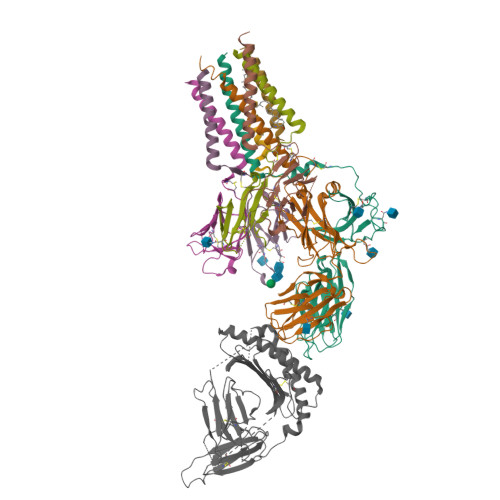
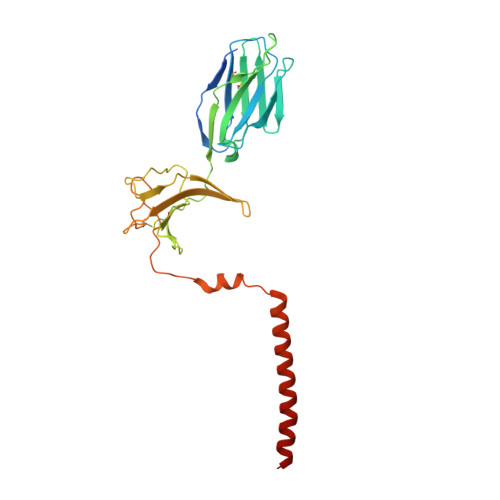
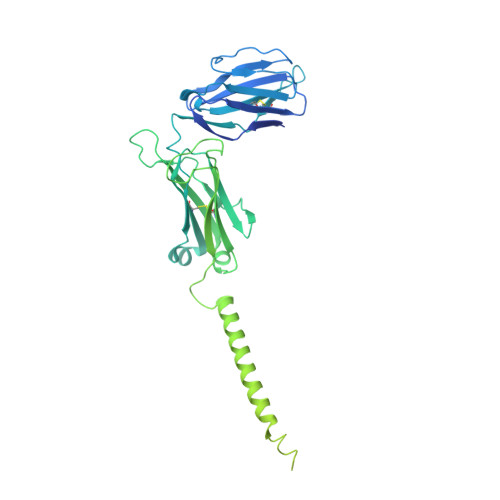
The resting and ligand-bound states of the membrane-embedded human T-cell receptor-CD3 complex.
Notti, R.Q., Yi, F., Heissel, S., Bush, M.W., Molvi, Z., Das, P., Molina, H., Klebanoff, C.A., Walz, T.(2024) bioRxiv
- PubMed: 37662363
- DOI: https://doi.org/10.1101/2023.08.22.554360
- Primary Citation of Related Structures:
8TW4, 8TW6, 9BBC, 9C3E - PubMed Abstract:
The T-cell receptor (TCR) initiates T-lymphocyte activation, but mechanistic questions remain( 1-4 ). Here, we present cryogenic electron microscopy structures for the unliganded and human leukocyte antigen (HLA)-bound human TCR-CD3 complex in nanodiscs that provide a native-like lipid environment. Distinct from the "open and extended" conformation seen in detergent( 5-8 ), the unliganded TCR-CD3 in nanodiscs adopts two related "closed and compacted" conformations that represent its physiologic resting state in vivo . By contrast, the HLA-bound complex adopts the open and extended conformation, and conformation-locking disulfide mutants show that ectodomain opening is necessary for maximal ligand-dependent T-cell activation. Together, these results reveal allosteric conformational change during TCR activation and highlight the importance of native-like lipid environments for membrane protein structure determination.