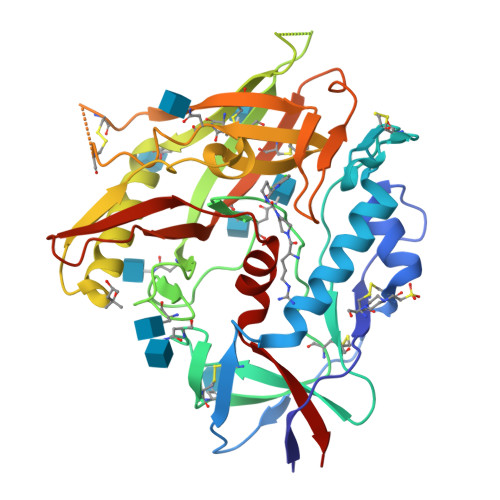
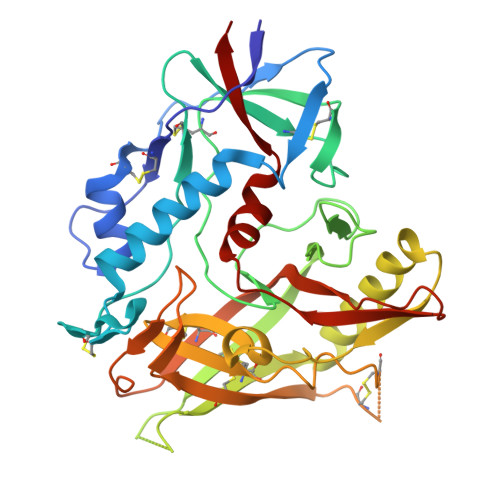
Optimization of a Piperidine CD4-Mimetic Scaffold Sensitizing HIV-1 Infected Cells to Antibody-Dependent Cellular Cytotoxicity.
Lee, D., Niu, L., Ding, S., Zhu, H., Tolbert, W.D., Medjahed, H., Beaudoin-Bussieres, G., Abrams, C., Finzi, A., Pazgier, M., Smith 3rd, A.B.(2024) ACS Med Chem Lett 15: 1961-1969
- PubMed: 39563795
- DOI: https://doi.org/10.1021/acsmedchemlett.4c00403
- Primary Citation of Related Structures:
9BXB, 9BXD, 9BXF, 9BXG, 9BXW, 9BXY - PubMed Abstract:
The ability of the HIV-1 accessory proteins Nef and Vpu to decrease CD4 protects infected cells from antibody-dependent cellular cytotoxicity (ADCC) by limiting the exposure of vulnerable epitopes to envelope glycoprotein (Env). Small-molecule CD4 mimetics (CD4mcs) based on piperidine scaffolds represent a new family of agents capable of sensitizing HIV-1-infected cells to ADCC by exposing CD4-induced (CD4i) epitopes on Env that are recognized by non-neutralizing antibodies which are abundant in plasma of people living with HIV. Here, we employed the combined methods of parallel synthesis, structure-based design, and optimization to generate a new line of piperidine-based CD4mcs, which sensitize HIV-1 infected cells to ADCC activity. The X-ray crystallographic study of the CD4mcs within the gp120 residues suggests that the positioning of the CD4mc inside the Phe43 cavity and synergistic contact of the CD4mc with the β 20-21 loop and the α 1 -helix lead to improved antiviral activity.
Organizational Affiliation:
Department of Chemistry, School of Arts and Sciences, University of Pennsylvania, Philadelphia, Pennsylvania 19104, United States.