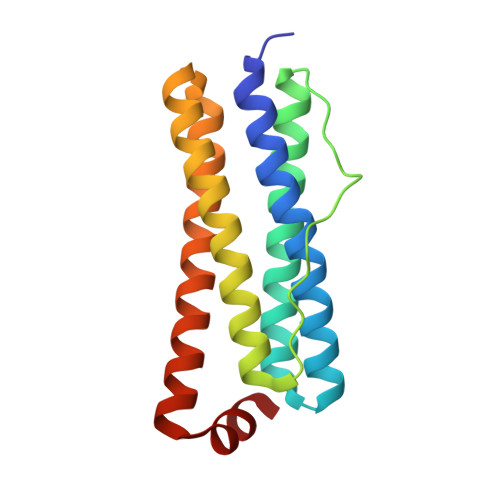
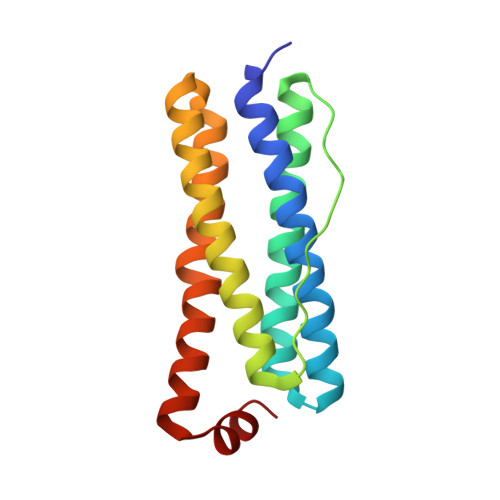
The crystal structure of Acinetobacter baumannii bacterioferritin reveals a heteropolymer of bacterioferritin and ferritin subunits.
Yao, H., Alli, S., Liu, L., Soldano, A., Cooper, A., Fontenot, L., Verdin, D., Battaile, K.P., Lovell, S., Rivera, M.(2024) Sci Rep 14: 18242-18242
- PubMed: 39107474
- DOI: https://doi.org/10.1038/s41598-024-69156-2
- Primary Citation of Related Structures:
9BTS - PubMed Abstract:
Iron storage proteins, e.g., vertebrate ferritin, and the ferritin-like bacterioferritin (Bfr) and bacterial ferritin (Ftn), are spherical, hollow proteins that catalyze the oxidation of Fe 2+ at binuclear iron ferroxidase centers (FOC) and store the Fe 3+ in their interior, thus protecting cells from unwanted Fe 3+ /Fe 2+ redox cycling and storing iron at concentrations far above the solubility of Fe 3+ . Vertebrate ferritins are heteropolymers of H and L subunits with only the H subunits having FOC. Bfr and Ftn were thought to coexist in bacteria as homopolymers, but recent evidence indicates these molecules are heteropolymers assembled from Bfr and Ftn subunits. Despite the heteropolymeric nature of vertebrate and bacterial ferritins, structures have been determined only for recombinant proteins constituted by a single subunit type. Herein we report the structure of Acinetobacter baumannii bacterioferritin, the first structural example of a heteropolymeric ferritin or ferritin-like molecule, assembled from completely overlapping Ftn homodimers harboring FOC and Bfr homodimers devoid of FOC but binding heme. The Ftn homodimers function by catalyzing the oxidation of Fe 2+ to Fe 3+ , while the Bfr homodimers bind a cognate ferredoxin (Bfd) which reduces the stored Fe 3+ by transferring electrons via the heme, enabling Fe 2+ mobilization to the cytosol for incorporation in metabolism.
Organizational Affiliation:
Department of Chemistry, Louisiana State University, Baton Rouge, 70803, USA.