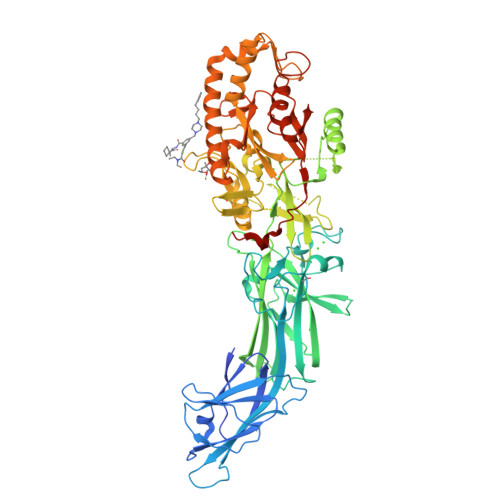
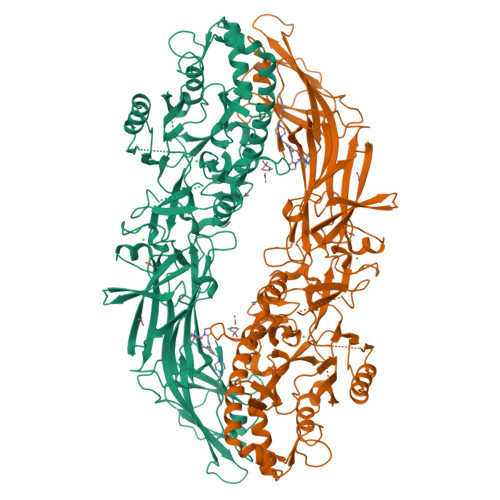
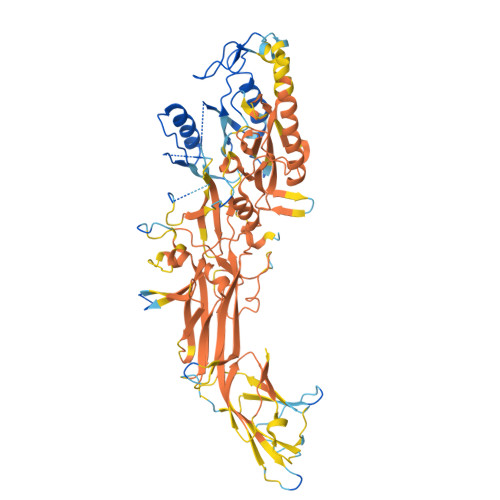
Discovery, Characterization, and Structure of a Cell Active PAD2 Inhibitor Acting through a Novel Allosteric Mechanism.
Byrnes, L.J., Choi, W.Y., Balbo, P., Banker, M.E., Chang, J., Chen, S., Cheng, X., Cong, Y., Culp, J., Di, H., Griffor, M., Hall, J., Meng, X., Morgan, B., Mousseau, J.J., Nicki, J., O'Connell, T., Ramsey, S., Shaginian, A., Shanker, S., Trujillo, J., Wan, J., Vincent, F., Wright, S.W., Vajdos, F.(2024) ACS Chem Biol 19: 2186-2197
- PubMed: 39316753
- DOI: https://doi.org/10.1021/acschembio.4c00397
- Primary Citation of Related Structures:
9B96, 9B97, 9B98 - PubMed Abstract:
Peptidyl arginine deiminases (PADs) are important enzymes in many diseases, especially those involving inflammation and autoimmunity. Despite many years of effort, developing isoform-specific inhibitors has been a challenge. We describe herein the discovery of a potent, noncovalent PAD2 inhibitor, with selectivity over PAD3 and PAD4, from a DNA-encoded library. The biochemical and biophysical characterization of this inhibitor and two noninhibitory binders indicated a novel, Ca 2+ competitive mechanism of inhibition. This was confirmed via X-ray crystallographic analysis. Finally, we demonstrate that this inhibitor selectively inhibits PAD2 in a cellular context.
Organizational Affiliation:
Pfizer Worldwide Research and Development, Eastern Pt. Rd, Groton, Connecticut 06340-5146, United States.