Proton-triggered rearrangement of the AMPA receptor N-terminal domains impacts receptor kinetics and synaptic localization
Ivica, J., Kejzar, N., Ho, H., Stockwell, I., Kuchtiak, V., Scrutton, A.M., Nakagawa, T., Greger, I.H.To be published.
Experimental Data Snapshot
Starting Models: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Voltage-dependent calcium channel gamma-2 subunit | A [auth H], B [auth F], G [auth E], H [auth G] | 323 | Mus musculus | Mutation(s): 0 Gene Names: Cacng2, Stg | 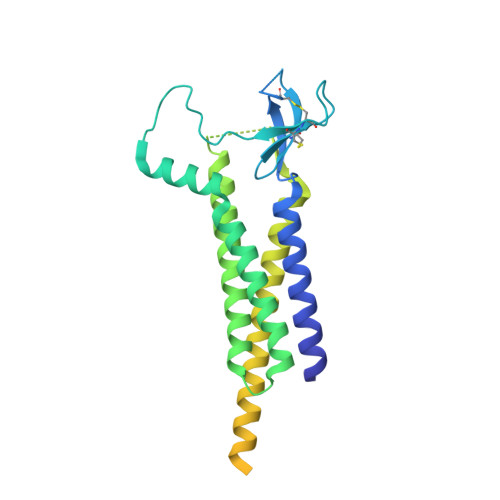 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for O88602 (Mus musculus) Explore O88602 Go to UniProtKB: O88602 | |||||
IMPC: MGI:1316660 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O88602 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Isoform Flip of Glutamate receptor 2 | C, D [auth B], E [auth A], F [auth D] | 889 | Rattus norvegicus | Mutation(s): 0 Gene Names: Gria2, Glur2 | 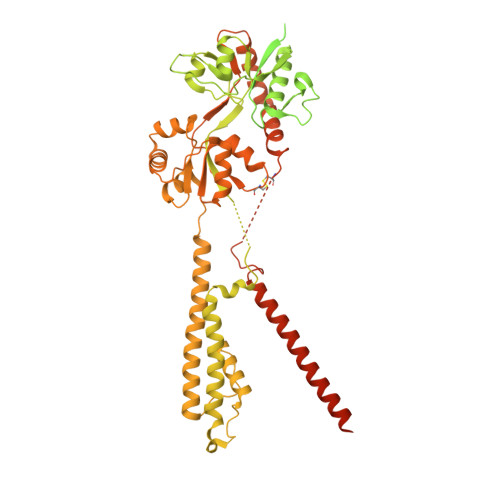 |
UniProt | |||||
Find proteins for P19491 (Rattus norvegicus) Go to UniProtKB: P19491 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P19491-2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.20.1_4487: |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of Mental Health (NIH/NIMH) | United States | R01MH123474 |
| UK Research and Innovation (UKRI) | United Kingdom | MC_U105174197 |
| Biotechnology and Biological Sciences Research Council (BBSRC) | United Kingdom | BB/N002113/1 |
| Wellcome Trust | United Kingdom | 223194/Z/21/Z |