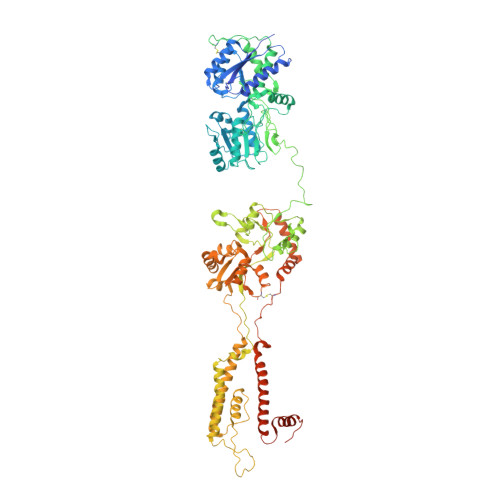
Kainate receptor channel opening and gating mechanism.
Gangwar, S.P., Yelshanskaya, M.V., Nadezhdin, K.D., Yen, L.Y., Newton, T.P., Aktolun, M., Kurnikova, M.G., Sobolevsky, A.I.(2024) Nature 630: 762-768
- PubMed: 38778115
- DOI: https://doi.org/10.1038/s41586-024-07475-0
- Primary Citation of Related Structures:
9B33, 9B34, 9B35, 9B36, 9B37, 9B38, 9B39 - PubMed Abstract:
Kainate receptors, a subclass of ionotropic glutamate receptors, are tetrameric ligand-gated ion channels that mediate excitatory neurotransmission 1-4 . Kainate receptors modulate neuronal circuits and synaptic plasticity during the development and function of the central nervous system and are implicated in various neurological and psychiatric diseases, including epilepsy, depression, schizophrenia, anxiety and autism 5-11 . Although structures of kainate receptor domains and subunit assemblies are available 12-18 , the mechanism of kainate receptor gating remains poorly understood. Here we present cryo-electron microscopy structures of the kainate receptor GluK2 in the presence of the agonist glutamate and the positive allosteric modulators lectin concanavalin A and BPAM344. Concanavalin A and BPAM344 inhibit kainate receptor desensitization and prolong activation by acting as a spacer between the amino-terminal and ligand-binding domains and a stabilizer of the ligand-binding domain dimer interface, respectively. Channel opening involves the kinking of all four pore-forming M3 helices. Our structures reveal the molecular basis of kainate receptor gating, which could guide the development of drugs for treatment of neurological disorders.
Organizational Affiliation:
Department of Biochemistry and Molecular Biophysics, Columbia University, New York, NY, USA.