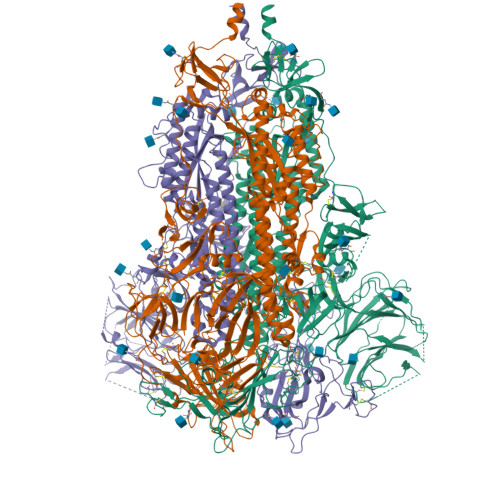
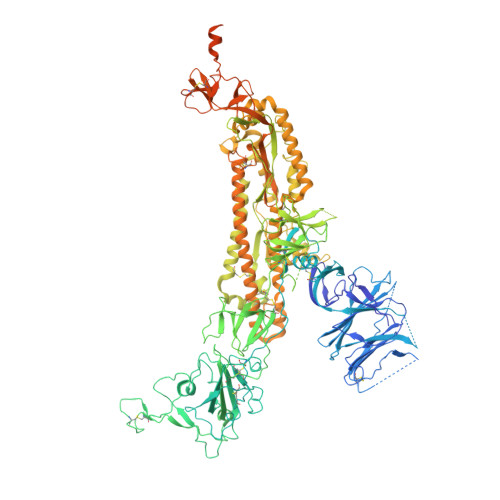
SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition.
Zhang, Q.E., Lindenberger, J., Parsons, R.J., Thakur, B., Parks, R., Park, C.S., Huang, X., Sammour, S., Janowska, K., Spence, T.N., Edwards, R.J., Martin, M., Williams, W.B., Gobeil, S., Montefiori, D.C., Korber, B., Saunders, K.O., Haynes, B.F., Henderson, R., Acharya, P.(2024) Mol Cell 84: 2747-2764.e7
- PubMed: 39059371
- DOI: https://doi.org/10.1016/j.molcel.2024.06.028
- Primary Citation of Related Structures:
8UIR, 8UK1, 8UKD, 8UKF, 8V0L, 8V0M, 8V0N, 8V0O, 8V0P, 8V0Q, 8V0R, 8V0S, 8V0T, 8V0U, 8V0V, 8V0W, 8V0X, 9AYW, 9AYX, 9AYY - PubMed Abstract:
A recombinant lineage of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron variant, named XBB, appeared in late 2022 and evolved descendants that successively swept local and global populations. XBB lineage members were noted for their improved immune evasion and transmissibility. Here, we determine cryoelectron microscopy (cryo-EM) structures of XBB.1.5, XBB.1.16, EG.5, and EG.5.1 spike (S) ectodomains to reveal reinforced 3-receptor binding domain (RBD)-down receptor-inaccessible closed states mediated by interprotomer RBD interactions previously observed in BA.1 and BA.2. Improved XBB.1.5 and XBB.1.16 RBD stability compensated for stability loss caused by early Omicron mutations, while the F456L substitution reduced EG.5 RBD stability. S1 subunit mutations had long-range impacts on conformation and epitope presentation in the S2 subunit. Our results reveal continued S protein evolution via simultaneous optimization of multiple parameters, including stability, receptor binding, and immune evasion, and the dramatic effects of relatively few residue substitutions in altering the S protein conformational landscape.
Organizational Affiliation:
Duke University, Duke Human Vaccine Institute, Durham, NC 27710, USA; Duke University, Department of Biochemistry, Durham, NC 27710, USA.