Structure-based engineering of monoclonal antibodies against fentanyl and carfentanil for improved binding
Rodarte, J.V., Pravetoni, M.To be published.
Experimental Data Snapshot
Starting Models: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| HY11-6B2_Mu Fab Heavy Chain | 228 | Mus musculus | Mutation(s): 0 | 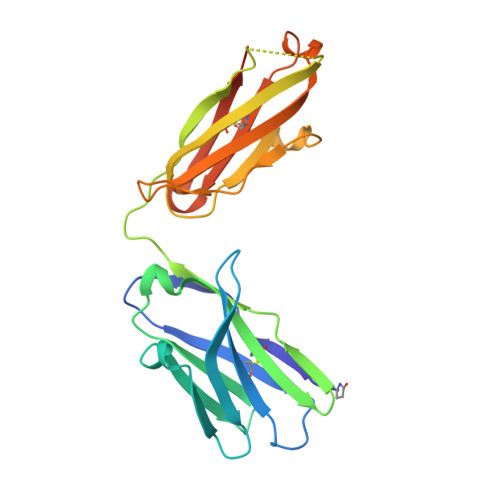 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| HY11-6B2_Mu Fab Light Chain | 214 | Mus musculus | Mutation(s): 0 | 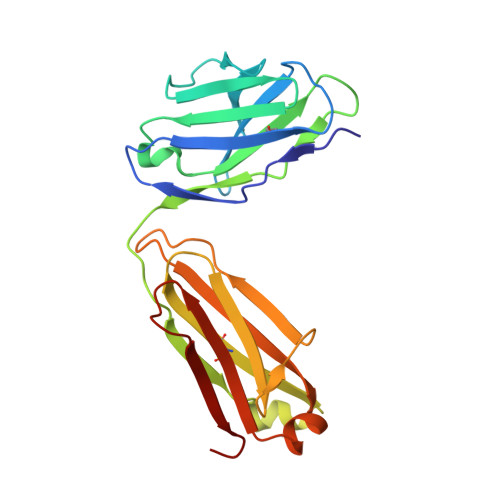 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| 7V7 (Subject of Investigation/LOI) Query on 7V7 | DB [auth O] GA [auth E] IB [auth R] Q [auth H] SA [auth I] | N-phenyl-N-[1-(2-phenylethyl)piperidin-4-yl]propanamide C22 H28 N2 O PJMPHNIQZUBGLI-UHFFFAOYSA-N |  | ||
| SO4 Query on SO4 | BA [auth D] CA [auth D] DA [auth D] EA [auth B] EB [auth O] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| EDO Query on EDO | AB [auth N] BB [auth N] CB [auth N] HB [auth Q] IA [auth E] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| NI Query on NI | AA [auth C], FA [auth B] | NICKEL (II) ION Ni VEQPNABPJHWNSG-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| PCA Query on PCA | A [auth H] C [auth A] D [auth C] G [auth E] J [auth I] A [auth H], C [auth A], D [auth C], G [auth E], J [auth I], K [auth M], M [auth O], O [auth Q] | L-PEPTIDE LINKING | C5 H7 N O3 |  | GLN |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 69.777 | α = 77.73 |
| b = 98.238 | β = 85.32 |
| c = 159.724 | γ = 77.95 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| Aimless | data scaling |
| HKL-3000 | data reduction |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute on Drug Abuse (NIH/NIDA) | United States | UG3 DA047711 |