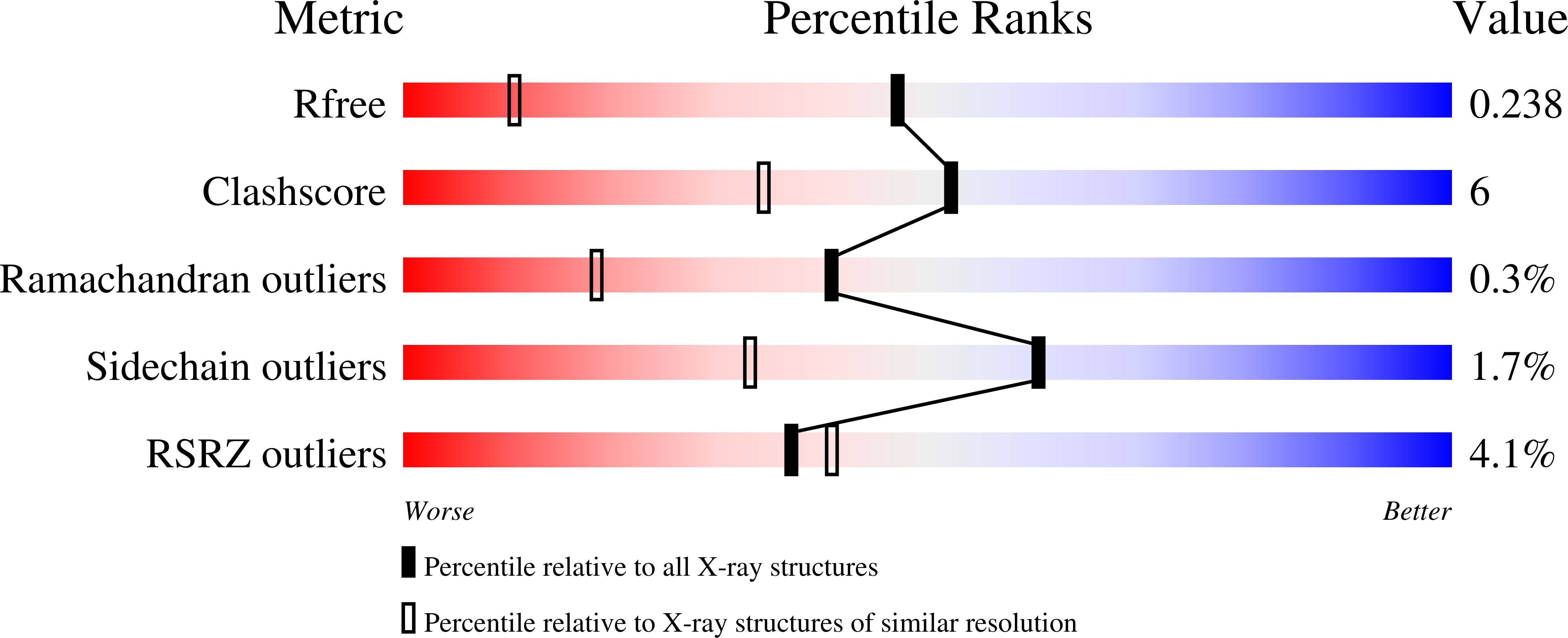
Structural and virologic mechanism of emergence of main protease inhibitor-resistance in SARS-CoV-2 as selected with main protease inhibitors
Hattori, S., Bulut, H., Hayashi, H., Kishimoto, N., Takamune, N., Hasegawa, K., Murayama, K., Mi, L., Wlodawer, A., Tamamura, H., Misumi, S., Mitsuya, H.To be published.