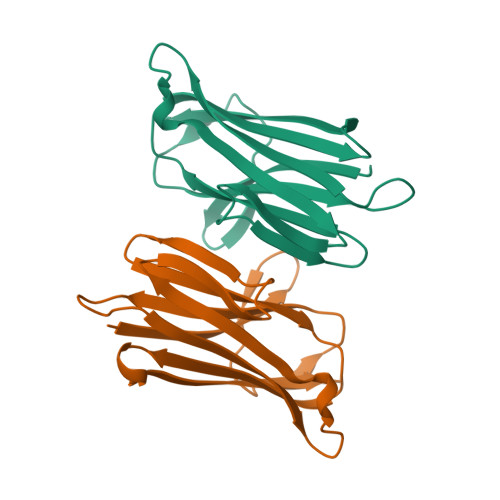
A large-scale type I CBASS antiphage screen identifies the phage prohead protease as a key determinant of immune activation and evasion.
Richmond-Buccola, D., Hobbs, S.J., Garcia, J.M., Toyoda, H., Gao, J., Shao, S., Lee, A.S.Y., Kranzusch, P.J.(2024) Cell Host Microbe 32: 1074-1088.e5
- PubMed: 38917809
- DOI: https://doi.org/10.1016/j.chom.2024.05.021
- Primary Citation of Related Structures:
9ARD - PubMed Abstract:
Cyclic oligonucleotide-based signaling system (CBASS) is an antiviral system that protects bacteria from phage infection and is evolutionarily related to human cGAS-STING immunity. cGAS-STING signaling is initiated by the recognition of viral DNA, but the molecular cues activating CBASS are incompletely understood. Using a screen of 975 type I CBASS operon-phage challenges, we show that operons with distinct cGAS/DncV-like nucleotidyltransferases (CD-NTases) and CD-NTase-associated protein (Cap) effectors exhibit marked patterns of phage restriction. We find that some type I CD-NTase enzymes require a C-terminal AGS-C immunoglobulin (Ig)-like fold domain for defense against select phages. Escaper phages evade CBASS via protein-coding mutations in virion assembly proteins, and acquired resistance is largely operon specific. We demonstrate that the phage Bas13 prohead protease interacts with the CD-NTase EcCdnD12 and can induce CBASS-dependent growth arrest in cells. Our results define phage virion assembly as a determinant of type I CBASS immune evasion and support viral protein recognition as a putative mechanism of cGAS-like enzyme activation.
Organizational Affiliation:
Department of Microbiology, Harvard Medical School, Boston, MA 02115, USA; Department of Cancer Immunology and Virology, Dana-Farber Cancer Institute, Boston, MA 02115, USA.