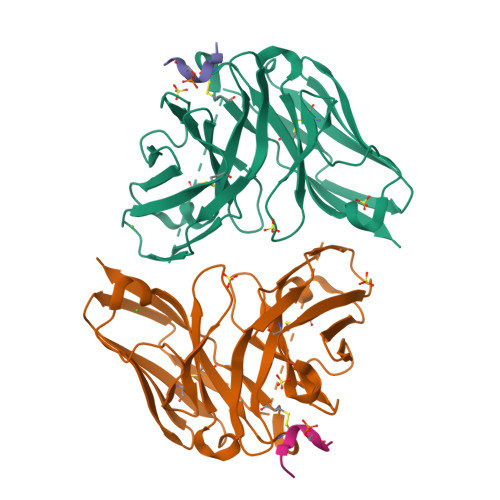
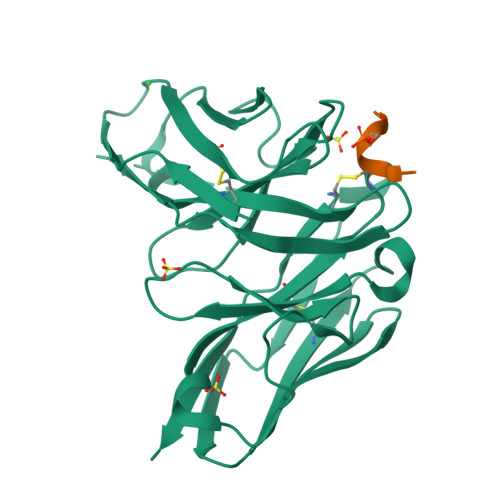
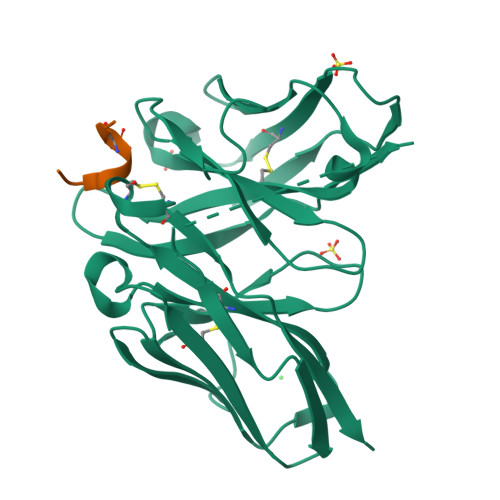
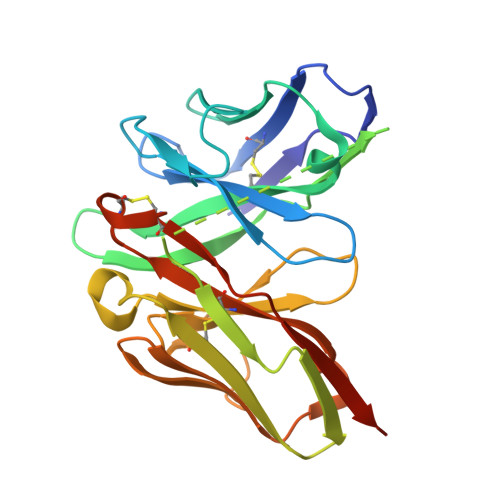
Unveiling the structural mechanisms behind high affinity and selectivity in phosphorylated epitope-specific rabbit antibodies.
Kasahara, K., Kawade, R., Nakakido, M., Matsunaga, R., Akiba, H., Entzminger, K.C., Maruyama, T., Okumura, S.C.J., Caaveiro, J.M.M., Kuroda, D., Tsumoto, K.(2024) J Biological Chem 300: 107989-107989
- PubMed: 39542251
- DOI: https://doi.org/10.1016/j.jbc.2024.107989
- Primary Citation of Related Structures:
8JOW, 8ZPU, 8ZXW - PubMed Abstract:
Protein phosphorylation is a crucial process in various cellular functions, and its irregularities have been implicated in several diseases, including cancer. Antibodies are commonly employed to detect protein phosphorylation in research. However, unlike the extensive studies on recognition mechanisms of the phosphate group by proteins such as kinases and phosphatases, only a few studies have explored antibody mechanisms. In this study, we produced and characterized two rabbit monoclonal antibodies that recognize a monophosphorylated Akt peptide. Through crystallography, thermodynamic mutational analyses, and molecular dynamics simulations, we investigated the unique recognition mechanism that enables higher binding affinity and selectivity of the antibodies compared to other generic proteins with lower binding affinity to phosphorylated epitopes. Our results demonstrate that molecular dynamics simulations provide novel insights into the dynamic aspects of molecular recognition of posttranslational modifications by proteins beyond static crystal structures, highlighting how specific atomic level interactions drive the exceptional affinity and selectivity of antibodies.
Organizational Affiliation:
Department of Bioengineering, School of Engineering, The University of Tokyo, Bunkyo-ku, Tokyo, Japan.