Molecular mechanism of prolactin-releasing peptide recognition and signaling via its G protein-coupled receptor.
Li, Y., Yuan, Q., He, X., Zhang, Y., You, C., Wu, C., Li, J., Xu, H.E., Zhao, L.H.(2024) Cell Discov 10: 91-91
- PubMed: 39223120
- DOI: https://doi.org/10.1038/s41421-024-00724-6
- Primary Citation of Related Structures:
8ZPS, 8ZPT - PubMed Abstract:
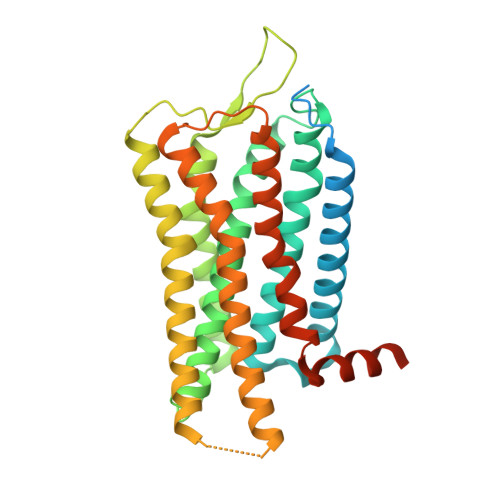
Prolactin-releasing peptide (PrRP) is an RF-amide neuropeptide that binds and activates its cognate G protein-coupled receptor, prolactin-releasing peptide receptor (PrRPR), also known as GPR10. PrRP and PrRPR are highly conserved across mammals and involved in regulating a range of physiological processes, including stress response, appetite regulation, pain modulation, cardiovascular function, and potentially reproductive functions. Here we present cryo-electron microscopy structures of PrRP-bound PrRPR coupled to G q or G i heterotrimer, unveiling distinct molecular determinants underlying the specific recognition of the ligand's C-terminal RF-amide motif. We identify a conserved polar pocket that accommodates the C-terminal amide shared by RF-amide peptides. Structural comparison with neuropeptide Y receptors reveals both similarities and differences in engaging the essential RF/RY-amide motifs. Our findings demonstrate the general mechanism governing RF-amide motif recognition by PrRPR and RF-amide peptide receptors, and provide a foundation for elucidating activation mechanisms and developing selective drugs targeting this important peptide-receptor system.
- State Key Laboratory of Drug Research, Center for Structure and Function of Drug Targets, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai, China.
Organizational Affiliation: