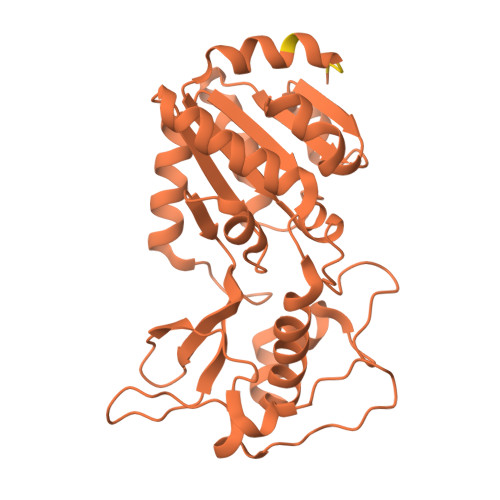
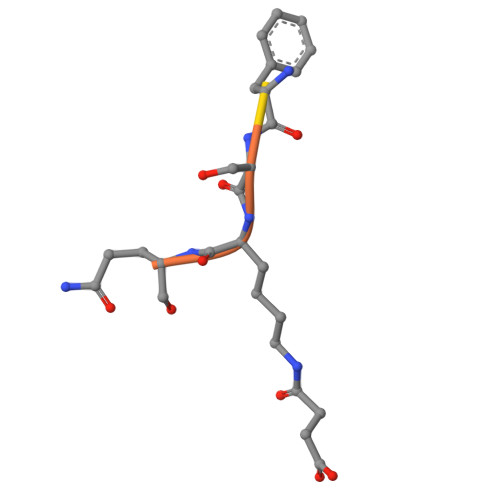
SIRT5 mutants reveal the role of conserved asparagine and glutamine residues in the NAD + -binding pocket.
Yokoyama, T., Takayama, Y., Mizuguchi, M., Nabeshima, Y., Kusaka, K.(2024) FEBS Lett 598: 2269-2280
- PubMed: 39031546
- DOI: https://doi.org/10.1002/1873-3468.14961
- Primary Citation of Related Structures:
8Z54, 8Z55, 8Z56, 8Z57, 8Z58 - PubMed Abstract:
SIRT5, one of the mammalian sirtuins, specifically recognizes succinyl-lysine residues on proteins and catalyzes the desuccinylation reaction. In this study, we characterized SIRT5 mutants with hydrophobic amino acid substitutions at Q140 and N141, in addition to the catalytic residue H158, known as an active site residue, by the Michaelis-Menten analysis and X-ray crystallography. Kinetic analysis showed that the catalytic efficiency (k cat /K m ) of the Q140L and N141V mutants decreased to 0.02 times and 0.0038 times that of the wild-type SIRT5, respectively, with the activity of the N141V mutant becoming comparable to that of the H158M mutant. Our findings indicate that N141 contributes significantly to the desuccinylation reaction.
- Faculty of Pharmaceutical Sciences, University of Toyama, Japan.
Organizational Affiliation: