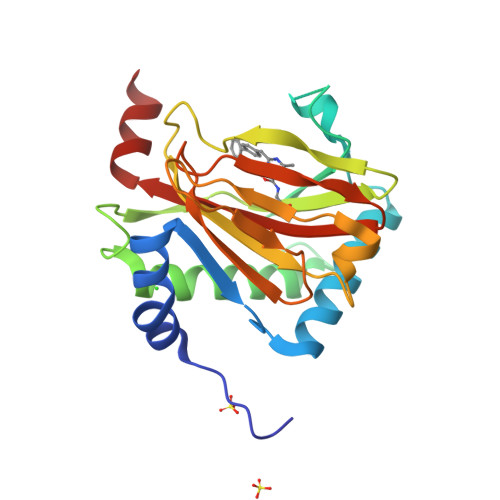
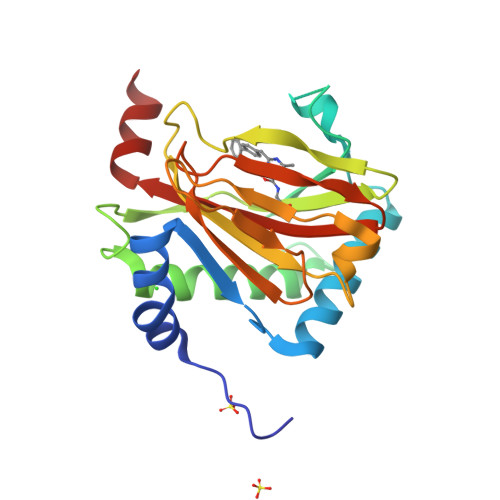
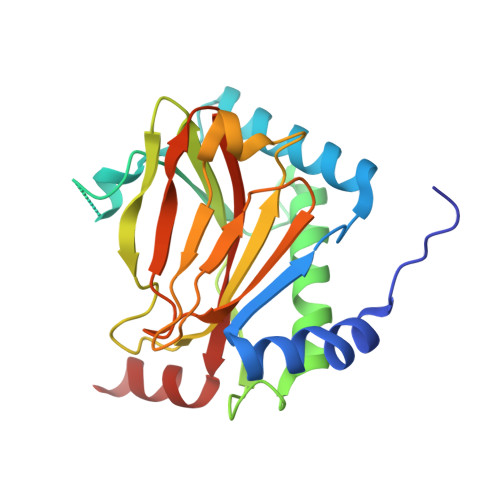
Discovery of DS44470011: An oral hypoxia-inducible factor prolyl hydroxylase inhibitor for the treatment of renal anemia.
Fukuda, T., Kuribayashi, T., Takano, R., Sasaki, K., Tsuji, T., Niitsu, Y., Ishii, K., Hashimoto, M., Baba, D., Ito, S., Tanaka, N.(2024) Bioorg Med Chem Lett 108: 129799-129799
- PubMed: 38754564
- DOI: https://doi.org/10.1016/j.bmcl.2024.129799
- Primary Citation of Related Structures:
8Z31, 8Z32, 8Z33, 8Z35 - PubMed Abstract:
Inhibition of the hypoxia-inducible factor prolyl hydroxylase (HIF-PHD) represents a promising strategy for discovering next-generation treatments for renal anemia. We identified a pyrimidine core with HIF-PHD inhibitory activity based on scaffold hopping of FG-2216 using crystal structures of HIF-PHD2 in complex with compound. By optimizing the substituents at the 2- and 6- positions of the pyrimidine core, we discovered DS44470011, which improves the effectiveness of erythropoietin (EPO) release in cells. Oral administration of DS44470011 to cynomolgus monkeys increased plasma EPO levels.
Organizational Affiliation:
R&D Division, Daiichi Sankyo Co., Ltd., 1-2-58 Hiromachi, Shinagawa-ku, Tokyo 140-8710, Japan. Electronic address: fukuda.takeshi.zv@daiichisankyo.co.jp.