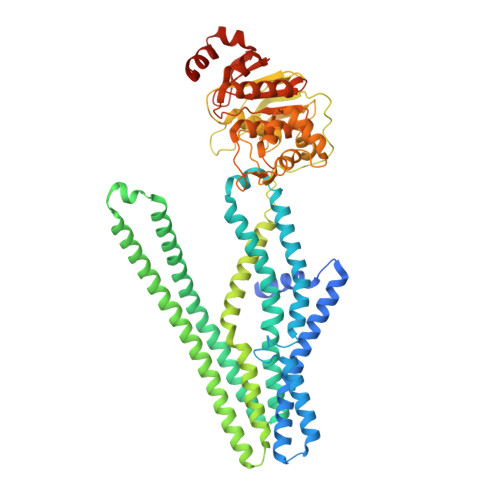
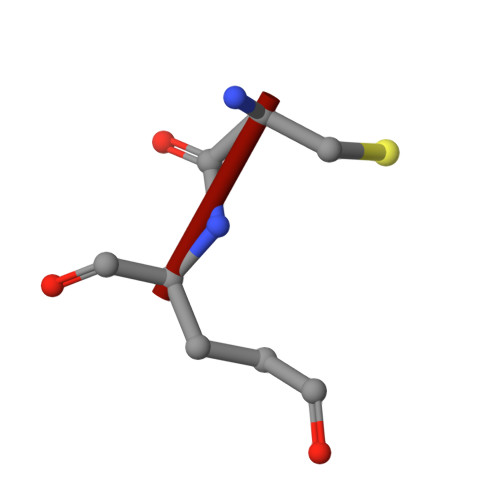
Cryo-EM structure of cadmium-bound human ABCB6.
Choi, S.H., Lee, S.S., Lee, H.Y., Kim, S., Kim, J.W., Jin, M.S.(2024) Commun Biol 7: 672-672
- PubMed: 38822018
- DOI: https://doi.org/10.1038/s42003-024-06377-1
- Primary Citation of Related Structures:
8YR3, 8YR4 - PubMed Abstract:
ATP-binding cassette transporter B6 (ABCB6), a protein essential for heme biosynthesis in mitochondria, also functions as a heavy metal efflux pump. Here, we present cryo-electron microscopy structures of human ABCB6 bound to a cadmium Cd(II) ion in the presence of antioxidant thiol peptides glutathione (GSH) and phytochelatin 2 (PC2) at resolutions of 3.2 and 3.1 Å, respectively. The overall folding of the two structures resembles the inward-facing apo state but with less separation between the two halves of the transporter. Two GSH molecules are symmetrically bound to the Cd(II) ion in a bent conformation, with the central cysteine protruding towards the metal. The N-terminal glutamate and C-terminal glycine of GSH do not directly interact with Cd(II) but contribute to neutralizing positive charges of the binding cavity by forming hydrogen bonds and van der Waals interactions with nearby residues. In the presence of PC2, Cd(II) binding to ABCB6 is similar to that observed with GSH, except that two cysteine residues of each PC2 molecule participate in Cd(II) coordination to form a tetrathiolate. Structural comparison of human ABCB6 and its homologous Atm-type transporters indicate that their distinct substrate specificity might be attributed to variations in the capping residues situated at the top of the substrate-binding cavity.
- School of Life Sciences, GIST, 123 Cheomdangwagi-ro, Buk-gu, Gwangju, Republic of Korea.
Organizational Affiliation: