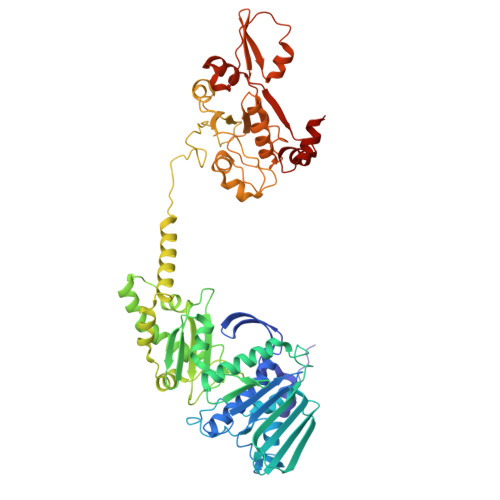
Structural and functional insights into the T-even type bacteriophage topoisomerase II.
Xin, Y., Xian, R., Yang, Y., Cong, J., Rao, Z., Li, X., Chen, Y.(2024) Nat Commun 15: 8719-8719
- PubMed: 39379365
- DOI: https://doi.org/10.1038/s41467-024-53037-3
- Primary Citation of Related Structures:
8YLU, 8YO1, 8YO3, 8YO4, 8YO5, 8YO7, 8YO9, 8YOD, 8YON, 9IMJ - PubMed Abstract:
T-even type bacteriophages are virulent phages commonly used as model organisms, playing a crucial role in understanding various biological processes. One such process involves the regulation of DNA topology during phage replication upon host infection, governed by type IIA DNA topoisomerases. In spite of various studies on prokaryotic and eukaryotic counterparts, viral topoisomerase II remains insufficiently understood, especially the unique domain composition of T4 phage. In this study, we determine the cryo-EM structures of topoisomerase II from T4 and T6 phages, including full-length structures of both apo and DNA-binding states which have never been determined before. Together with other conformational states, these structures provide an explicit blueprint of mechanisms of phage topoisomerase II. Particularly, the asymmetric dimeric interactions observed in cryo-EM structures of T6 phage topoisomerase II ATPase domain and central domain bound with DNA shed light on the asynchronous ATP usage and asynchronous cleavage of the G-segment DNA, respectively. The elucidation of phage topoisomerase II's structures and functions not only enhances our understanding of mechanisms and evolutionary parallels with prokaryotic and eukaryotic homologs but also highlights its potential as a model for developing type IIA topoisomerase inhibitors.
- National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing, China.
Organizational Affiliation: