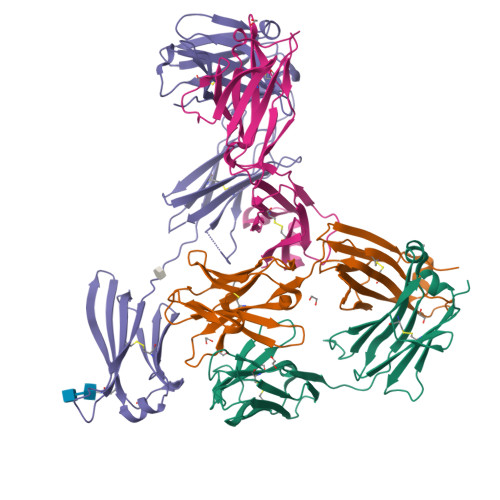
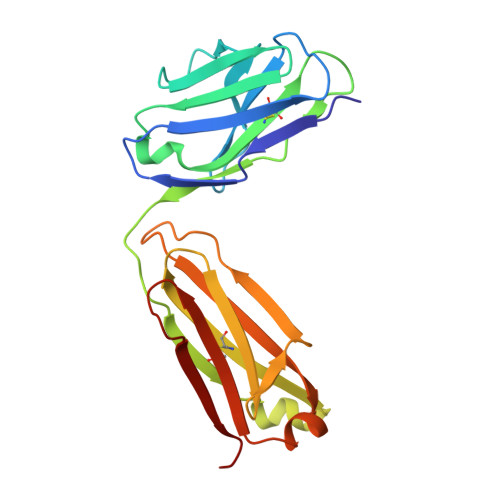
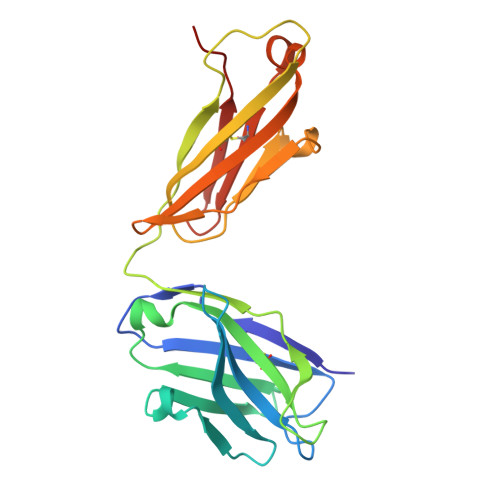
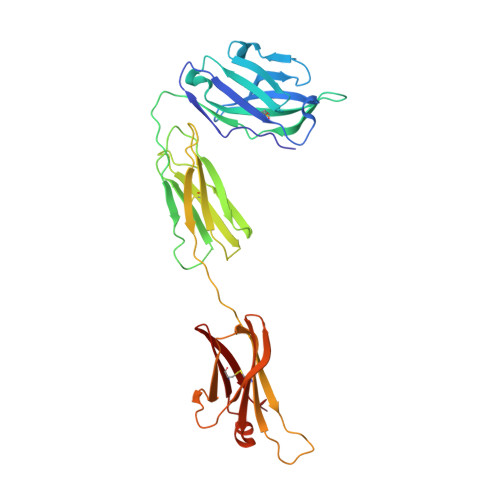
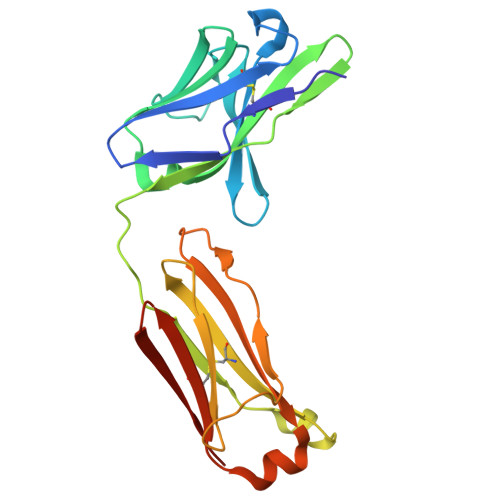
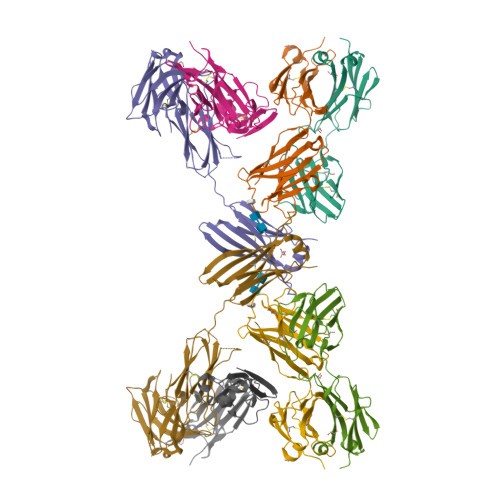Allosteric inhibition of IgE-Fc epsilon RI interactions by simultaneous targeting of IgE F(ab')2 epitopes.
Hirano, T., Koyanagi, A., Ago, H., Yamamoto, M., Kitaura, J., Kasai, M., Okumura, K.(2024) Commun Biol 7: 1042-1042
- PubMed: 39179708
- DOI: https://doi.org/10.1038/s42003-024-06633-4
- Primary Citation of Related Structures:
8XX0 - PubMed Abstract:
Immunoglobulin E (IgE) plays pivotal roles in allergic diseases through interaction with a high-affinity receptor (FcεRI). We established that Fab fragments of anti-IgE antibodies (HMK-12 Fab) rapidly dissociate preformed IgE-FcεRI complexes in a temperature-dependent manner and inhibit IgE-mediated anaphylactic reactions, even after allergen challenge. X-ray crystallographic studies revealed that HMK-12 Fab interacts with each of two equivalent epitopes on the Cε2 homodimer domain involved in IgE F(ab')2. Consequently, HMK-12 Fab-mediated targeting of Cε2 reduced the binding affinity of Fc domains and resulted in rapid removal of IgE from the receptor complex. This unexpected finding of allosteric inhibition of IgE-FcεRI interactions by simultaneous targeting of two epitope sites on the Cε2 homodimer domain of IgE F(ab')2 may have implications for the development of novel therapies for allergic disease.
Organizational Affiliation:
Department of Hematology, Juntendo University Nerima Hospital, Nerima-ku, Tokyo, Japan. thirano@juntendo.ac.jp.