Optimization of Novel Pyrido-pyridazinone Derivatives as FER Tyrosine Kinase Inhibitors, Leading to the Potent DS08701581.
Taniguchi, T., Yasumatsu, I., Inagaki, H., Baba, D., Toyota, A., Kaneta, Y., Odagiri, T., Momose, T., Kawai, J., Imaoka, T., Nakayama, K.(2024) ACS Med Chem Lett 15: 1010-1016
- PubMed: 39015278
- DOI: https://doi.org/10.1021/acsmedchemlett.4c00030
- Primary Citation of Related Structures:
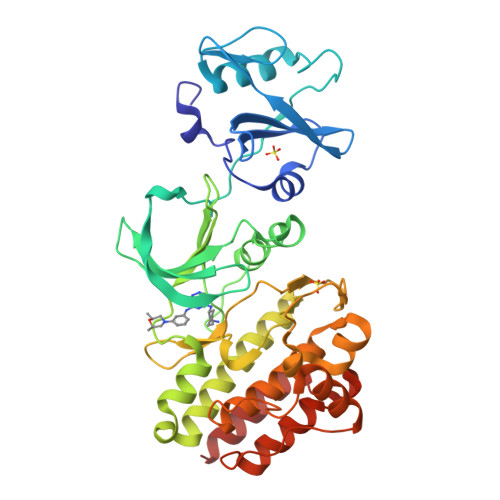
8XKP - PubMed Abstract:
Previously, we reported the new pyrido-pyridazinone template as a feline sarcoma-related (FER) tyrosine kinase inhibitor. Representative compound 1 ( DS21360717 ) showed strong enzyme inhibitory activity (IC50 = 0.5 nM), however, its antitumor effect was insufficient, probably due to poor solubility and resultant low bioavailability (BA). In addition, the kinase selectivity was inadequate, which may result in certain safety risks. Here, we focused on derivatization of the unoptimized C-5 position to obtain promising FER inhibitors possessing strong antitumor effects and improved selectivity, referring to their X-ray crystal structure and the docking model with FES proto-oncogene tyrosine kinase as an FER surrogate. While establishing the synthetic route of the pyrido-pyridazinone scaffold, we obtained a desired compound via our derivatization. Our optimized compound 17c (DS08701581 ) showed the highest class cell-free and cell activities in this template, good oral BA, and improved kinase selectivity, resulting in significant tumor growth inhibition in the Ba/F3-FER tumor model without body weight loss.
Organizational Affiliation:
R&D Division, Daiichi Sankyo Co., Ltd., 1-2-58 Hiromachi, Shinagawa-ku, Tokyo 140-8710, Japan.