Cryo-EM structure of integrin ITGAV, ITGB3 and CYCLO (RGDFK) complex, conformation 2
Xi, Z.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Integrin alpha-V | 1,048 | Homo sapiens | Mutation(s): 0 Gene Names: ITGAV | 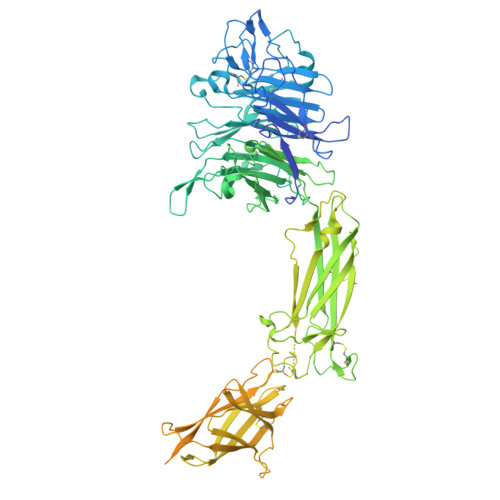 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P06756 (Homo sapiens) Explore P06756 Go to UniProtKB: P06756 | |||||
PHAROS: P06756 GTEx: ENSG00000138448 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P06756 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Integrin beta-3 | 788 | Homo sapiens | Mutation(s): 0 Gene Names: ITGB3 |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P05106 (Homo sapiens) Explore P05106 Go to UniProtKB: P05106 | |||||
PHAROS: P05106 GTEx: ENSG00000259207 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P05106 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| A1LU7 Query on A1LU7 | C [auth B] | 2-[(2~{R},5~{R},8~{S},11~{S})-8-(4-azanylbutyl)-11-(3-carbamimidamidopropyl)-3,6,9,12,15-pentakis(oxidanylidene)-5-(phenylmethyl)-1,4,7,10,13-pentazacyclopentadec-2-yl]ethanoic acid C27 H41 N9 O7 NVHPXYIRNJFKTE-VNTMZGSJSA-N |  | ||
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | -- |