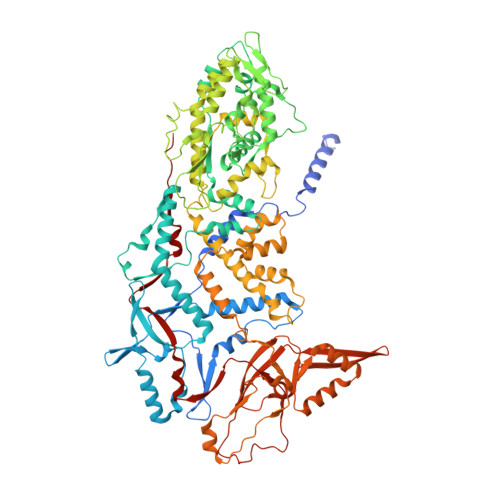
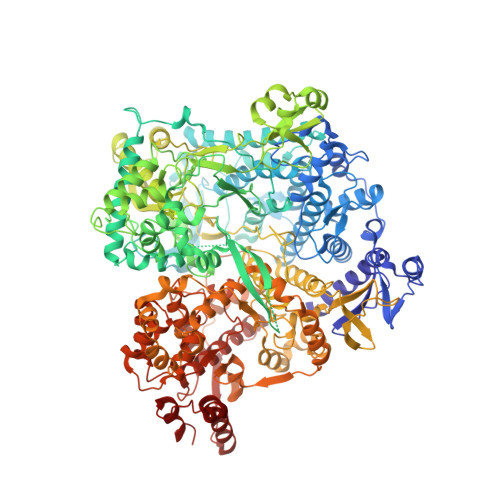
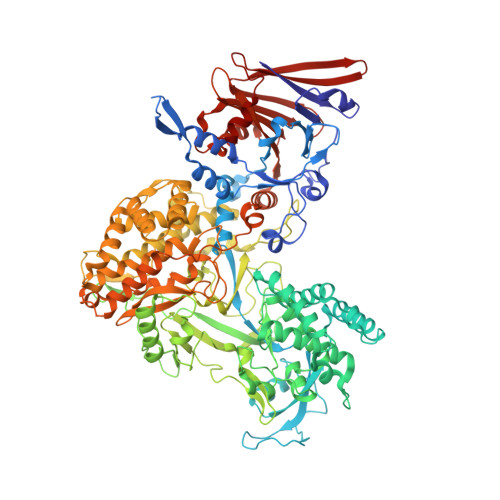
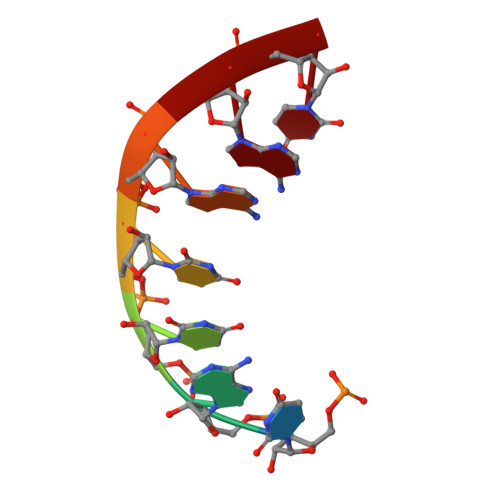
RNA genome packaging and capsid assembly of bluetongue virus visualized in host cells.
Xia, X., Sung, P.Y., Martynowycz, M.W., Gonen, T., Roy, P., Zhou, Z.H.(2024) Cell 187: 2236-2249.e17
- PubMed: 38614100
- DOI: https://doi.org/10.1016/j.cell.2024.03.007
- Primary Citation of Related Structures:
8W12, 8W19, 8W1C, 8W1I, 8W1O, 8W1R, 8W1S - PubMed Abstract:
Unlike those of double-stranded DNA (dsDNA), single-stranded DNA (ssDNA), and ssRNA viruses, the mechanism of genome packaging of dsRNA viruses is poorly understood. Here, we combined the techniques of high-resolution cryoelectron microscopy (cryo-EM), cellular cryoelectron tomography (cryo-ET), and structure-guided mutagenesis to investigate genome packaging and capsid assembly of bluetongue virus (BTV), a member of the Reoviridae family of dsRNA viruses. A total of eleven assembly states of BTV capsid were captured, with resolutions up to 2.8 Å, with most visualized in the host cytoplasm. ATPase VP6 was found underneath the vertices of capsid shell protein VP3 as an RNA-harboring pentamer, facilitating RNA packaging. RNA packaging expands the VP3 shell, which then engages middle- and outer-layer proteins to generate infectious virions. These revealed "duality" characteristics of the BTV assembly mechanism reconcile previous contradictory co-assembly and core-filling models and provide insights into the mysterious RNA packaging and capsid assembly of Reoviridae members and beyond.
- Department of Microbiology, Immunology and Molecular Genetics, University of California, Los Angeles, Los Angeles, CA 90095, USA; California NanoSystems Institute, University of California, Los Angeles, Los Angeles, CA 90095, USA.
Organizational Affiliation: