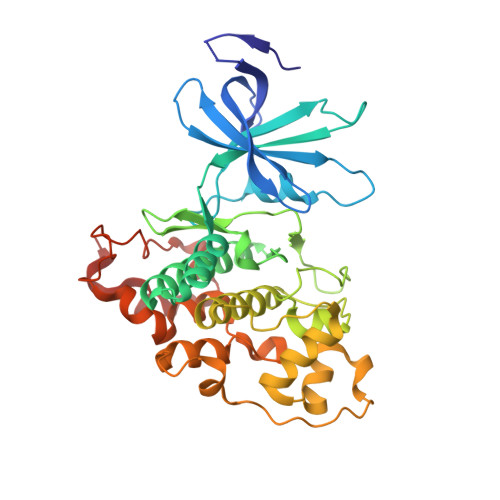
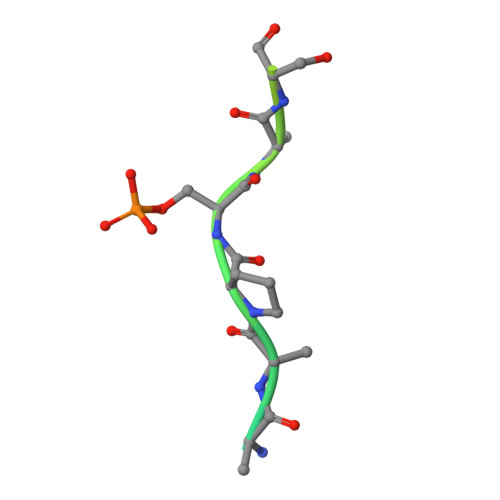
Structural and functional effects of phosphopriming and scaffolding in the kinase GSK-3 beta.
Enos, M.D., Gavagan, M., Jameson, N., Zalatan, J.G., Weis, W.I.(2024) Sci Signal 17
- PubMed: 39226374
- DOI: https://doi.org/10.1126/scisignal.ado0881
- Primary Citation of Related Structures:
8VME, 8VMF, 8VMG - PubMed Abstract:
Glycogen synthase kinase 3β (GSK-3β) targets specific signaling pathways in response to distinct upstream signals. We used structural and functional studies to dissect how an upstream phosphorylation step primes the Wnt signaling component β-catenin for phosphorylation by GSK-3β and how scaffolding interactions contribute to this reaction. Our crystal structure of GSK-3β bound to a phosphoprimed β-catenin peptide confirmed the expected binding mode of the phosphoprimed residue adjacent to the catalytic site. An aspartate phosphomimic in the priming site of β-catenin adopted an indistinguishable structure but reacted approximately 1000-fold slower than the native phosphoprimed substrate. This result suggests that substrate positioning alone is not sufficient for catalysis and that native phosphopriming interactions are necessary. We also obtained a structure of GSK-3β with an extended peptide from the scaffold protein Axin that bound with greater affinity than that of previously crystallized Axin fragments. This structure neither revealed additional contacts that produce the higher affinity nor explained how substrate interactions in the GSK-3β active site are modulated by remote Axin binding. Together, our findings suggest that phosphopriming and scaffolding produce small conformational changes or allosteric effects, not captured in the crystal structures, that activate GSK-3β and facilitate β-catenin phosphorylation. These results highlight limitations in our ability to predict catalytic activity from structure and have potential implications for the role of natural phosphomimic mutations in kinase regulation and phosphosite evolution.
- Department of Structural Biology, Stanford University School of Medicine, Stanford, CA 94035, USA.
Organizational Affiliation: