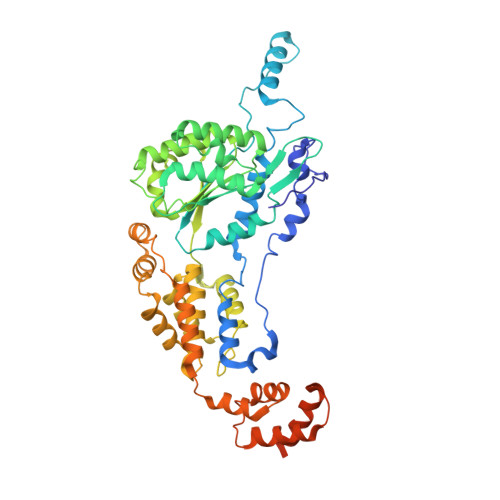
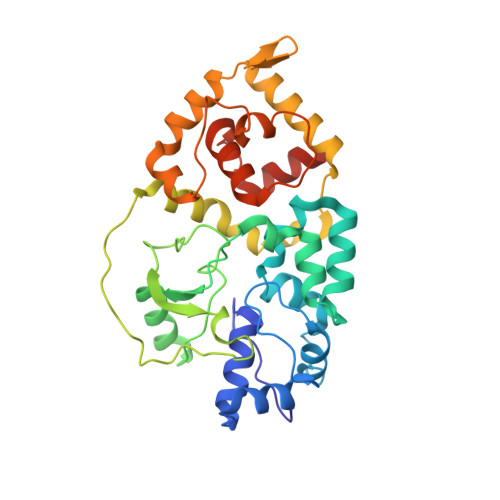
Assembly of the Tn7 targeting complex by a regulated stepwise process.
Shen, Y., Krishnan, S.S., Petassi, M.T., Hancock, M.A., Peters, J.E., Guarne, A.(2024) Mol Cell 84: 2368-2381.e6
- PubMed: 38834067
- DOI: https://doi.org/10.1016/j.molcel.2024.05.012
- Primary Citation of Related Structures:
8GLU, 8GLW, 8GLX, 8VCJ, 8VCT - PubMed Abstract:
The Tn7 family of transposons is notable for its highly regulated integration mechanisms, including programmable RNA-guided transposition. The targeting pathways rely on dedicated target selection proteins from the TniQ family and the AAA+ adaptor TnsC to recruit and activate the transposase at specific target sites. Here, we report the cryoelectron microscopy (cryo-EM) structures of TnsC bound to the TniQ domain of TnsD from prototypical Tn7 and unveil key regulatory steps stemming from unique behaviors of ATP- versus ADP-bound TnsC. We show that TnsD recruits ADP-bound dimers of TnsC and acts as an exchange factor to release one protomer with exchange to ATP. This loading process explains how TnsC assembles a heptameric ring unidirectionally from the target site. This unique loading process results in functionally distinct TnsC protomers within the ring, providing a checkpoint for target immunity and explaining how insertions at programmed sites precisely occur in a specific orientation across Tn7 elements.
- Department of Biochemistry, McGill University, Montreal, QC H3G 0B1, Canada; Centre de recherche en biologie structurale (CRBS), McGill University, Montreal, QC H3G 0B1, Canada.
Organizational Affiliation: