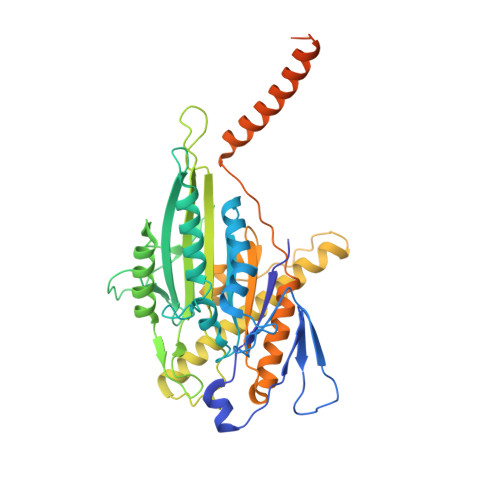
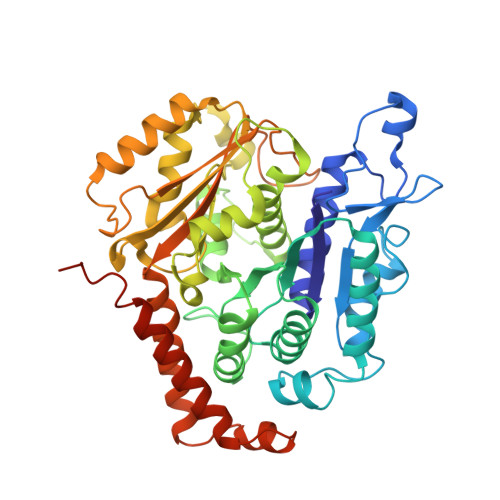
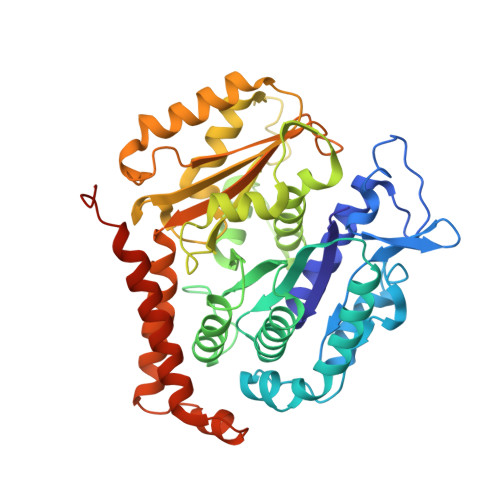
Cryo-EM unveils kinesin KIF1A's processivity mechanism and the impact of its pathogenic variant P305L.
Benoit, M.P.M.H., Rao, L., Asenjo, A.B., Gennerich, A., Sosa, H.(2024) Nat Commun 15: 5530-5530
- PubMed: 38956021
- DOI: https://doi.org/10.1038/s41467-024-48720-4
- Primary Citation of Related Structures:
8UTN, 8UTO, 8UTP, 8UTQ, 8UTR, 8UTS, 8UTT, 8UTU, 8UTV, 8UTW, 8UTY - PubMed Abstract:
Mutations in the microtubule-associated motor protein KIF1A lead to severe neurological conditions known as KIF1A-associated neurological disorders (KAND). Despite insights into its molecular mechanism, high-resolution structures of KIF1A-microtubule complexes remain undefined. Here, we present 2.7-3.5 Å resolution structures of dimeric microtubule-bound KIF1A, including the pathogenic P305L mutant, across various nucleotide states. Our structures reveal that KIF1A binds microtubules in one- and two-heads-bound configurations, with both heads exhibiting distinct conformations with tight inter-head connection. Notably, KIF1A's class-specific loop 12 (K-loop) forms electrostatic interactions with the C-terminal tails of both α- and β-tubulin. The P305L mutation does not disrupt these interactions but alters loop-12's conformation, impairing strong microtubule-binding. Structure-function analysis reveals the K-loop and head-head coordination as major determinants of KIF1A's superprocessive motility. Our findings advance the understanding of KIF1A's molecular mechanism and provide a basis for developing structure-guided therapeutics against KAND.
- Department of Biochemistry and Gruss-Lipper Biophotonics Center, Albert Einstein College of Medicine, Bronx, NY, 10461, USA. matthieu.benoit@univ-rennes.fr.
Organizational Affiliation: